An Introduction to Machine Learning
- PMID: 32128792
- PMCID: PMC7189875
- DOI: 10.1002/cpt.1796
An Introduction to Machine Learning
Abstract
In the last few years, machine learning (ML) and artificial intelligence have seen a new wave of publicity fueled by the huge and ever-increasing amount of data and computational power as well as the discovery of improved learning algorithms. However, the idea of a computer learning some abstract concept from data and applying them to yet unseen situations is not new and has been around at least since the 1950s. Many of these basic principles are very familiar to the pharmacometrics and clinical pharmacology community. In this paper, we want to introduce the foundational ideas of ML to this community such that readers obtain the essential tools they need to understand publications on the topic. Although we will not go into the very details and theoretical background, we aim to point readers to relevant literature and put applications of ML in molecular biology as well as the fields of pharmacometrics and clinical pharmacology into perspective.
© 2020 The Authors. Clinical Pharmacology & Therapeutics published by Wiley Periodicals, Inc. on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
All authors declared no competing interests for this work.
Figures
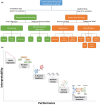

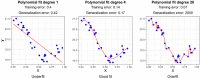
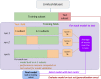


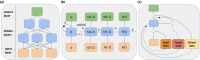
References
-
- Camacho, D.M. , Collins, K.M. , Powers, R.K. , Costello, J.C. & Collins, J.J. Next‐generation machine learning for biological networks. Cell 173, 1581–1592 (2018). - PubMed
-
- Rajkomar, A. , Dean, J. & Kohane, I. Machine learning in medicine. N. Engl. J. Med. 380, 1347–1358 (2019). - PubMed
-
- Kleene, S.C. Representation of Events in Nerve Nets and Finite Automata (RAND Project Air Force, Santa Monica, CA, 1951) <https://apps.dtic.mil/docs/citations/ADA596138>.
-
- Breiman, L. Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat. Sci. 16, 199–231 (2001).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

