Proteome profile of peripheral myelin in healthy mice and in a neuropathy model
- PMID: 32130108
- PMCID: PMC7056269
- DOI: 10.7554/eLife.51406
Proteome profile of peripheral myelin in healthy mice and in a neuropathy model
Abstract
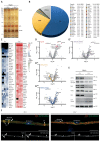
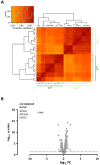
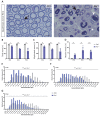
Proteome and transcriptome analyses aim at comprehending the molecular profiles of the brain, its cell-types and subcellular compartments including myelin. Despite the relevance of the peripheral nervous system for normal sensory and motor capabilities, analogous approaches to peripheral nerves and peripheral myelin have fallen behind evolving technical standards. Here we assess the peripheral myelin proteome by gel-free, label-free mass-spectrometry for deep quantitative coverage. Integration with RNA-Sequencing-based developmental mRNA-abundance profiles and neuropathy disease genes illustrates the utility of this resource. Notably, the periaxin-deficient mouse model of the neuropathy Charcot-Marie-Tooth 4F displays a highly pathological myelin proteome profile, exemplified by the discovery of reduced levels of the monocarboxylate transporter MCT1/SLC16A1 as a novel facet of the neuropathology. This work provides the most comprehensive proteome resource thus far to approach development, function and pathology of peripheral myelin, and a straightforward, accurate and sensitive workflow to address myelin diversity in health and disease.
Keywords: charcot-marie-tooth (cmt) neuropathy; demyelination; mouse; myelin proteome; neuroscience; peripheral nervous system; schwann cell.
© 2020, Siems et al.
Conflict of interest statement
SS, OJ, ME, NK, LW, DS, KK, DH, RJ, RF, MS, MR, PB, HW No competing interests declared
Figures



References
-
- Ackerman SD, Luo R, Poitelon Y, Mogha A, Harty BL, D'Rozario M, Sanchez NE, Lakkaraju AKK, Gamble P, Li J, Qu J, MacEwan MR, Ray WZ, Aguzzi A, Feltri ML, Piao X, Monk KR. GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. Journal of Experimental Medicine. 2018;215:941–961. doi: 10.1084/jem.20161714. - DOI - PMC - PubMed
-
- Ambrozkiewicz MC, Schwark M, Kishimoto-Suga M, Borisova E, Hori K, Salazar-Lázaro A, Rusanova A, Altas B, Piepkorn L, Bessa P, Schaub T, Zhang X, Rabe T, Ripamonti S, Rosário M, Akiyama H, Jahn O, Kobayashi T, Hoshino M, Tarabykin V, Kawabe H. Polarity acquisition in cortical neurons is driven by synergistic action of Sox9-Regulated Wwp1 and Wwp2 E3 ubiquitin ligases and intronic miR-140. Neuron. 2018;100:1097–1115. doi: 10.1016/j.neuron.2018.10.008. - DOI - PubMed

