Hierarchy in Hfq Chaperon Occupancy of Small RNA Targets Plays a Major Role in Their Regulation
- PMID: 32130912
- PMCID: PMC7059120
- DOI: 10.1016/j.celrep.2020.02.016
Hierarchy in Hfq Chaperon Occupancy of Small RNA Targets Plays a Major Role in Their Regulation
Abstract
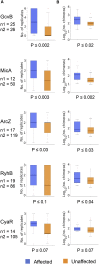
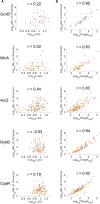
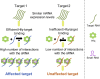
Bacterial small RNAs (sRNAs) are posttranscriptional regulators of gene expression that base pair with complementary sequences on target mRNAs, often in association with the chaperone Hfq. Here, using experimentally identified sRNA-target pairs, along with gene expression measurements, we assess basic principles of regulation by sRNAs. We show that the sRNA sequence dictates the target repertoire, as point mutations in the sRNA shift the target set correspondingly. We distinguish two subsets of targets: targets showing changes in expression levels under overexpression of their sRNA regulator and unaffected targets that interact more sporadically with the sRNA. These differences among targets are associated with their Hfq occupancy, rather than with the sRNA-target base-pairing potential. Our results suggest that competition among targets over Hfq binding plays a major role in the regulatory outcome, possibly awarding targets with higher Hfq binding efficiency an advantage in the competition over binding to the sRNA.
Keywords: Hfq; RIL-seq; RNA sequencing; posttranscriptional regulation; small RNAs.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
-
- Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

