NUDT7 Loss Promotes KrasG12D CRC Development
- PMID: 32131398
- PMCID: PMC7139971
- DOI: 10.3390/cancers12030576
NUDT7 Loss Promotes KrasG12D CRC Development
Abstract
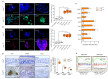
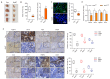
Studies have suggested that dysregulation of peroxisomal lipid metabolism might play an important role in colorectal cancer (CRC) development. Here, we found that KrasG12D-driven CRC tumors demonstrate dysfunctional peroxisomal b-oxidation and identified Nudt7 (peroxisomal coenzyme A diphosphatase NUDT7) as one of responsible peroxisomal genes. In KrasG12D-driven CRC tumors, the expression level of Nudt7 was significantly decreased. Treatment of azoxymethane/dextran sulfate sodium (AOM/DSS) into Nudt7 knockout (Nudt7-/-) mice significantly induced lipid accumulation and the expression levels of CRC-related genes whereas xenografting of Nudt7-overexpressed LS-174T cells into mice significantly reduced lipid accumulation and the expression levels of CRC-related genes. Ingenuity pathway analysis of microarray using the colon of Nudt7-/- and Nudt7+/+ mice treated with AOM/DSS suggested Wnt signaling as one of activated signaling pathways in Nudt7-/- colons. Upregulated levels of β-catenin were observed in the colons of KrasG12D and AOM/DSS-treated Nudt7-/- mice and downstream targets of β-catenin such as Myc, Ccdn1, and Nos2, were also significantly increased in the colon of Nudt7-/- mice. We observed an increased level of palmitic acid in the colon of Nudt7-/- mice and attachment of palmitic acid-conjugated chitosan patch into the colon of mice induced the expression levels of b-catenin and CRC-related genes. Overall, our data reveal a novel role for peroxisomal NUDT7 in KrasG12D-driven CRC development.
Keywords: colorectal cancer; palmitic acid; peroxisomal coenzyme A diphosphatase NUDT7 (NUDT7); peroxisome; β-catenin.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures




References
-
- Bhandari A., Woodhouse M., Gupta S. Colorectal cancer is a leading cause of cancer incidence and mortality among adults younger than 50 years in the USA: A SEER-based analysis with comparison to other young-onset cancers. J. Investig. Med. 2017;65:311–315. doi: 10.1136/jim-2016-000229. - DOI - PMC - PubMed
-
- Gylfe A.E., Katainen R., Kondelin J., Tanskanen T., Cajuso T., Hanninen U., Taipale J., Taipale M., Renkonen-Sinisalo L., Jarvinen H., et al. Eleven candidate susceptibility genes for common familial colorectal cancer. PLoS Genet. 2013;9:e1003876. doi: 10.1371/journal.pgen.1003876. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

