Identifying Novel ATX Inhibitors via Combinatory Virtual Screening Using Crystallography-Derived Pharmacophore Modelling, Docking Study, and QSAR Analysis
- PMID: 32131468
- PMCID: PMC7179221
- DOI: 10.3390/molecules25051107
Identifying Novel ATX Inhibitors via Combinatory Virtual Screening Using Crystallography-Derived Pharmacophore Modelling, Docking Study, and QSAR Analysis
Abstract
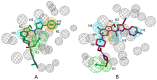
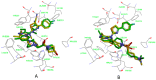
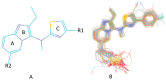
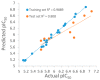
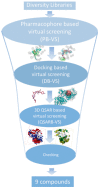
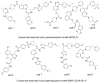
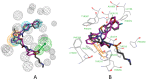
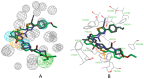
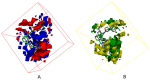
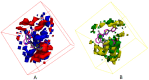
Autotaxin (ATX) is considered as an interesting drug target for the therapy of several diseases. The goal of the research was to detect new ATX inhibitors which have novel scaffolds by using virtual screening. First, based on two diverse receptor-ligand complexes, 14 pharmacophore models were developed, and the 14 models were verified through a big test database. Those pharmacophore models were utilized to accomplish virtual screening. Next, for the purpose of predicting the probable binding poses of compounds and then carrying out further virtual screening, docking-based virtual screening was performed. Moreover, an excellent 3D QSAR model was established, and 3D QSAR-based virtual screening was applied for predicting the activity values of compounds which got through the above two-round screenings. A correlation coefficient r2, which equals 0.988, was supplied by the 3D QSAR model for the training set, and the correlation coefficient r2 equaling 0.808 for the test set means that the developed 3D QSAR model is an excellent model. After the filtering was done by the combinatory virtual screening, which is based on the pharmacophore modelling, docking study, and 3D QSAR modelling, we chose nine potent inhibitors with novel scaffolds finally. Furthermore, two potent compounds have been particularly discussed.
Keywords: 3D QSAR model; autotaxin inhibitor; docking calculation; pharmacophore model; virtual screening.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
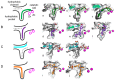


Similar articles
-
In silico approaches to identify novel myeloid cell leukemia-1 (Mcl-1) inhibitors for treatment of cancer.J Biomol Struct Dyn. 2018 Jul;36(9):2424-2435. doi: 10.1080/07391102.2017.1356241. Epub 2017 Aug 22. J Biomol Struct Dyn. 2018. PMID: 28714799
-
Identification of novel VP35 inhibitors: Virtual screening driven new scaffolds.Biomed Pharmacother. 2016 Dec;84:199-207. doi: 10.1016/j.biopha.2016.09.034. Epub 2016 Sep 19. Biomed Pharmacother. 2016. PMID: 27657828
-
Pharmacophore Based 3D-QSAR, Virtual Screening and Docking Studies on Novel Series of HDAC Inhibitors with Thiophen Linker as Anticancer Agents.Comb Chem High Throughput Screen. 2016;19(9):735-751. doi: 10.2174/1386207319666160801154415. Comb Chem High Throughput Screen. 2016. PMID: 27487787
-
Identification of Potent Small-Molecule PCSK9 Inhibitors Based on Quantitative Structure-Activity Relationship, Pharmacophore Modeling, and Molecular Docking Procedure.Curr Probl Cardiol. 2023 Jun;48(6):101660. doi: 10.1016/j.cpcardiol.2023.101660. Epub 2023 Feb 24. Curr Probl Cardiol. 2023. PMID: 36841313 Review.
-
Detection of Natural Inhibitors against Human Liver Cancer Cell Lines through QSAR, Molecular Docking and ADMET Studies.Curr Top Med Chem. 2021;21(8):686-695. doi: 10.2174/1568026620666201204155830. Curr Top Med Chem. 2021. PMID: 33280598 Review.
Cited by
-
Design and Development of Autotaxin Inhibitors.Pharmaceuticals (Basel). 2021 Nov 22;14(11):1203. doi: 10.3390/ph14111203. Pharmaceuticals (Basel). 2021. PMID: 34832985 Free PMC article. Review.
-
Discovery of novel JAK1 inhibitors through combining machine learning, structure-based pharmacophore modeling and bio-evaluation.J Transl Med. 2023 Aug 28;21(1):579. doi: 10.1186/s12967-023-04443-6. J Transl Med. 2023. PMID: 37641144 Free PMC article.
-
Integration of Ligand-Based and Structure-Based Methods for the Design of Small-Molecule TLR7 Antagonists.Molecules. 2022 Jun 23;27(13):4026. doi: 10.3390/molecules27134026. Molecules. 2022. PMID: 35807273 Free PMC article.
-
Computational Approaches to Molecular Properties, Chemical Reactivity, and Drug Virtual Screening.Molecules. 2020 Nov 13;25(22):5301. doi: 10.3390/molecules25225301. Molecules. 2020. PMID: 33202912 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

