Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status
- PMID: 32131884
- PMCID: PMC7057466
- DOI: 10.1186/s13062-020-0258-5
Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status
Abstract
Background: Accurate classification of different Burkholderia cepacia complex (BCC) species is essential for therapy, prognosis assessment and research. The taxonomic status of BCC remains problematic and an improved knowledge about the classification of BCC is in particular needed.
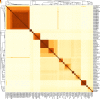
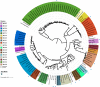
Methods: We compared phylogenetic trees of BCC based on 16S rRNA, recA, hisA and MLSA (multilocus sequence analysis). Using the available whole genome sequences of BCC, we inferred a species tree based on estimated single-copy orthologous genes and demarcated species of BCC using dDDH/ANI clustering.
Results: We showed that 16S rRNA, recA, hisA and MLSA have limited resolutions in the taxonomic study of closely related bacteria such as BCC. Our estimated species tree and dDDH/ANI clustering clearly separated 116 BCC strains into 36 clusters. With the appropriate reclassification of misidentified strains, these clusters corresponded to 22 known species as well as 14 putative novel species.
Conclusions: This is the first large-scale and systematic study of the taxonomic status of the BCC and could contribute to further insights into BCC taxonomy. Our study suggested that conjunctive use of core phylogeny based on single-copy orthologous genes, as well as pangenome-based dDDH/ANI clustering would provide a preferable framework for demarcating closely related species.
Reviewer: This article was reviewed by Dr. Xianwen Ren.
Keywords: Burkholderia cepacia complex; Classification; Genome; Single-copy orthologous genes; Taxonomy; dDDH and ANI.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Bach E, Sant'Anna FH, Magrich dos Passos JF, Balsanelli E, de Baura VA, Pedrosa FO, et al. Detection of misidentifications of species from the Burkholderia cepacia complex and description of a new member, the soil bacterium Burkholderia catarinensis sp. nov. Pathog Dis. 2017;75:6. - PubMed
-
- Laraya-Cuasay LR, Lipstein M, Huang NN. Pseudomonas cepacia in the respiratory flora of patients with cystic fibrosis (CF) Pediatr Res. 1977;11(4):502. doi: 10.1203/00006450-197704000-00792. - DOI
-
- Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, et al. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47(4):1188–1200. doi: 10.1099/00207713-47-4-1188. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

