Twitching and Swimming Motility Play a Role in Ralstonia solanacearum Pathogenicity
- PMID: 32132161
- PMCID: PMC7056806
- DOI: 10.1128/mSphere.00740-19
Twitching and Swimming Motility Play a Role in Ralstonia solanacearum Pathogenicity
Abstract
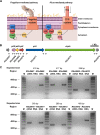
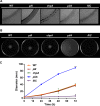
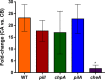
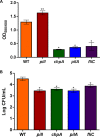
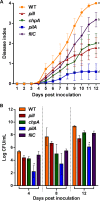
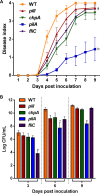
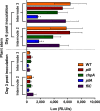
Ralstonia solanacearum is a bacterial plant pathogen causing important economic losses worldwide. In addition to the polar flagella responsible for swimming motility, this pathogen produces type IV pili (TFP) that govern twitching motility, a flagellum-independent movement on solid surfaces. The implication of chemotaxis in plant colonization, through the control flagellar rotation by the proteins CheW and CheA, has been previously reported in R. solanacearum In this work, we have identified in this bacterium homologues of the Pseudomonas aeruginosapilI and chpA genes, suggested to play roles in TFP-associated motility analogous to those played by the cheW and cheA genes, respectively. We demonstrate that R. solanacearum strains with a deletion of the pilI or the chpA coding region show normal swimming and chemotaxis but altered biofilm formation and reduced twitching motility, transformation efficiency, and root attachment. Furthermore, these mutants displayed wild-type growth in planta and impaired virulence on tomato plants after soil-drench inoculations but not when directly applied to the xylem. Comparison with deletion mutants for pilA and fliC-encoding the major pilin and flagellin subunits, respectively-showed that both twitching and swimming are required for plant colonization and full virulence. This work proves for the first time the functionality of a pilus-mediated pathway encoded by pil-chp genes in R. solanacearum, demonstrating that pilI and chpA genes are bona fide motility regulators controlling twitching motility and its three related phenotypes: virulence, natural transformation, and biofilm formation.IMPORTANCE Twitching and swimming are two bacterial movements governed by pili and flagella. The present work identifies for the first time in the Gram-negative plant pathogen Ralstonia solanacearum a pilus-mediated chemotaxis pathway analogous to that governing flagellum-mediated chemotaxis. We show that regulatory genes in this pathway control all of the phenotypes related to pili, including twitching motility, natural transformation, and biofilm formation, and are also directly implicated in virulence, mainly during the first steps of the plant infection. Our results show that pili have a higher impact than flagella on the interaction of R. solanacearum with tomato plants and reveal new types of cross-talk between the swimming and twitching motility phenotypes: enhanced swimming in bacteria lacking pili and a role for the flagellum in root attachment.
Keywords: Ralstonia solanacearum; chpA; fliC; pilA; pilI.
Copyright © 2020 Corral et al.
Figures
References
-
- Allen C, Prior P, Hayward AC. 2005. Bacterial wilt disease and the Ralstonia solanacearum species complex. American Phytopathological Society Press, St. Paul, MN.

