TDP-43 α-helical structure tunes liquid-liquid phase separation and function
- PMID: 32132204
- PMCID: PMC7084079
- DOI: 10.1073/pnas.1912055117
TDP-43 α-helical structure tunes liquid-liquid phase separation and function
Abstract
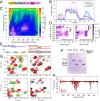
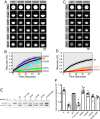
Liquid-liquid phase separation (LLPS) is involved in the formation of membraneless organelles (MLOs) associated with RNA processing. The RNA-binding protein TDP-43 is present in several MLOs, undergoes LLPS, and has been linked to the pathogenesis of amyotrophic lateral sclerosis (ALS). While some ALS-associated mutations in TDP-43 disrupt self-interaction and function, here we show that designed single mutations can enhance TDP-43 assembly and function via modulating helical structure. Using molecular simulation and NMR spectroscopy, we observe large structural changes upon dimerization of TDP-43. Two conserved glycine residues (G335 and G338) are potent inhibitors of helical extension and helix-helix interaction, which are removed in part by variants at these positions, including the ALS-associated G335D. Substitution to helix-enhancing alanine at either of these positions dramatically enhances phase separation in vitro and decreases fluidity of phase-separated TDP-43 reporter compartments in cells. Furthermore, G335A increases TDP-43 splicing function in a minigene assay. Therefore, the TDP-43 helical region serves as a short but uniquely tunable module where application of biophysical principles can precisely control assembly and function in cellular and synthetic biology applications of LLPS.
Keywords: NMR spectroscopy; liquid–liquid phase separation; molecular simulation; protein interactions.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Brangwynne C., Hoege C., Gharakhani J., Jülicher F., Hyman A. A., Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009). - PubMed
-
- Hyman A. A., Weber C. A., Jülicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

