Clonal tracking using embedded viral barcoding and high-throughput sequencing
- PMID: 32132718
- PMCID: PMC7427513
- DOI: 10.1038/s41596-019-0290-z
Clonal tracking using embedded viral barcoding and high-throughput sequencing
Abstract
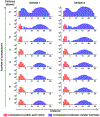
Embedded viral barcoding in combination with high-throughput sequencing is a powerful technology with which to track single-cell clones. It can provide clonal-level insights into cellular proliferation, development, differentiation, migration, and treatment efficacy. Here, we present a detailed protocol for a viral barcoding procedure that includes the creation of barcode libraries, the viral delivery of barcodes, the recovery of barcodes, and the computational analysis of barcode sequencing data. The entire procedure can be completed within a few weeks. This barcoding method requires cells to be susceptible to viral transduction. It provides high sensitivity and throughput, and enables precise quantification of cellular progeny. It is cost efficient and does not require any advanced skills. It can also be easily adapted to many types of applications, including both in vitro and in vivo experiments.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures



References
-
- Dick JE, Magli MC, Huszar D, Phillips RA & Bernstein A Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell 42, 71–79 (1985). - PubMed
-
- Keller G, Paige C, Gilboa E & Wagner EF Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature 318, 149–154 (1985). - PubMed
-
- Lemischka IR, Raulet DH & Mulligan RC Developmental potential and dynamic behavior of hematopoietic stem cells. Cell 45, 917–927 (1986). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R00 HL113104/HL/NHLBI NIH HHS/United States
- R35 HL150826/HL/NHLBI NIH HHS/United States
- R01HL135292S/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- R01HL135292/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- R01 HL135292/HL/NHLBI NIH HHS/United States
- P30 CA014089/CA/NCI NIH HHS/United States
- R00HL113104/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- R01 HL138225/HL/NHLBI NIH HHS/United States
- R01HL138225/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

