Development and validation of a risk prediction model to diagnose Barrett's oesophagus (MARK-BE): a case-control machine learning approach
- PMID: 32133440
- PMCID: PMC7056359
- DOI: 10.1016/S2589-7500(19)30216-X
Development and validation of a risk prediction model to diagnose Barrett's oesophagus (MARK-BE): a case-control machine learning approach
Abstract
Background: Screening for Barrett's Oesophagus (BE) relies on endoscopy which is invasive and has a low yield. This study aimed to develop and externally validate a simple symptom and risk-factor questionnaire to screen for patients with BE.
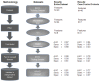
Methods: Questionnaires from 1299 patients in the BEST2 case-controlled study were analysed: 880 had BE including 40 with invasive oesophageal adenocarcinoma (OAC) and 419 were controls. This was randomly split into a training cohort of 776 patients and an internal validation cohort of 523 patients. External validation included 398 patients from the BOOST case-controlled study: 198 with BE (23 with OAC) and 200 controls. Identification of independently important diagnostic features was undertaken using machine learning techniques information gain (IG) and correlation based feature selection (CFS). Multiple classification tools were assessed to create a multi-variable risk prediction model. Internal validation was followed by external validation in the independent dataset.
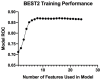
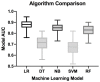
Findings: The BEST2 study included 40 features. Of these, 24 added IG but following CFS, only 8 demonstrated independent diagnostic value including age, gender, smoking, waist circumference, frequency of stomach pain, duration of heartburn and acid taste and taking of acid suppression medicines. Logistic regression offered the highest prediction quality with AUC (area under the receiver operator curve) of 0.87. In the internal validation set, AUC was 0.86. In the BOOST external validation set, AUC was 0.81.
Interpretation: The diagnostic model offers valid predictions of diagnosis of BE in patients with symptomatic gastroesophageal reflux, assisting in identifying who should go forward to invasive testing. Overweight men who have been taking stomach medicines for a long time may merit particular consideration for further testing. The risk prediction tool is quick and simple to administer but will need further calibration and validation in a prospective study in primary care.
Funding: Charles Wolfson Trust and Guts UK.
Conflict of interest statement
Declarations of Interest The Cytosponge device was designed by RCF and her research team in between 2009 and 2010. Patents and a trademark were filed in 2010 by the Medical Research Council (MRC). The BEST2 study was designed in 2010 and the device was manufactured for the specific purpose of this study following a letter of no objection from the Medical Health Regulatory Agency. In 2013 the MRC licensed the technology to Covidien GI Solutions, now part of Medtronic Inc. They have had no influence in any way on the design, conduct or analysis of this study. RCF, is a named inventor on patents pertaining to the Cytosponge and related assays. She has not received any financial benefits to date. All other authors have no conflicts of interest to declare
Figures

Comment in
-
Finding Barrett's oesophagus: is there a machine learning approach in our future?Lancet Digit Health. 2020 Jan;2(1):e6-e7. doi: 10.1016/S2589-7500(19)30226-2. Epub 2019 Dec 23. Lancet Digit Health. 2020. PMID: 33328039 No abstract available.
References
-
- Ross-Innes CSCSCS, Debiram-Beecham I, Walker E, et al. Evaluation of a Minimally Invasive Cell Sampling Device Coupled with Assessment of Trefoil Factor 3 Expression for Diagnosing Barrett’s Esophagus: A Multi-Center Case–Control Study. PLoS Med. 2015;12(1):1–19. doi: 10.1371/journal.pmed.1001780. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
