Mutation signatures specific to DNA alkylating agents in yeast and cancers
- PMID: 32133535
- PMCID: PMC7144945
- DOI: 10.1093/nar/gkaa150
Mutation signatures specific to DNA alkylating agents in yeast and cancers
Abstract
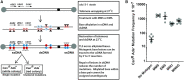
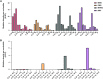
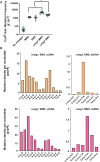
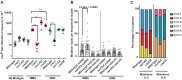
Alkylation is one of the most ubiquitous forms of DNA lesions. However, the motif preferences and substrates for the activity of the major types of alkylating agents defined by their nucleophilic substitution reactions (SN1 and SN2) are still unclear. Utilizing yeast strains engineered for large-scale production of single-stranded DNA (ssDNA), we probed the substrate specificity, mutation spectra and signatures associated with DNA alkylating agents. We determined that SN1-type agents preferably mutagenize double-stranded DNA (dsDNA), and the mutation signature characteristic of the activity of SN1-type agents was conserved across yeast, mice and human cancers. Conversely, SN2-type agents preferably mutagenize ssDNA in yeast. Moreover, the spectra and signatures derived from yeast were detectable in lung cancers, head and neck cancers and tumors from patients exposed to SN2-type alkylating chemicals. The estimates of mutation loads associated with the SN2-type alkylation signature were higher in lung tumors from smokers than never-smokers, pointing toward the mutagenic activity of the SN2-type alkylating carcinogens in cigarettes. In summary, our analysis of mutations in yeast strains treated with alkylating agents, as well as in whole-exome and whole-genome-sequenced tumors identified signatures highly specific to alkylation mutagenesis and indicate the pervasive nature of alkylation-induced mutagenesis in cancers.
Published by Oxford University Press on behalf of Nucleic Acids Research 2020.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

