Brain aging comprises many modes of structural and functional change with distinct genetic and biophysical associations
- PMID: 32134384
- PMCID: PMC7162660
- DOI: 10.7554/eLife.52677
Brain aging comprises many modes of structural and functional change with distinct genetic and biophysical associations
Abstract
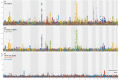
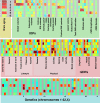
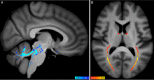
Brain imaging can be used to study how individuals' brains are aging, compared against population norms. This can inform on aspects of brain health; for example, smoking and blood pressure can be seen to accelerate brain aging. Typically, a single 'brain age' is estimated per subject, whereas here we identified 62 modes of subject variability, from 21,407 subjects' multimodal brain imaging data in UK Biobank. The modes represent different aspects of brain aging, showing distinct patterns of functional and structural brain change, and distinct patterns of association with genetics, lifestyle, cognition, physical measures and disease. While conventional brain-age modelling found no genetic associations, 34 modes had genetic associations. We suggest that it is important not to treat brain aging as a single homogeneous process, and that modelling of distinct patterns of structural and functional change will reveal more biologically meaningful markers of brain aging in health and disease.
Keywords: UK Biobank; brain aging; brain imaging; human; neuroscience.
© 2020, Smith et al.
Conflict of interest statement
SS, LE, FA, PM, TN, GD, KM No competing interests declared
Figures
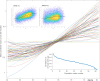




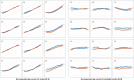
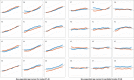
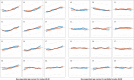


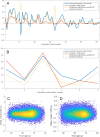
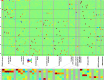
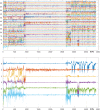
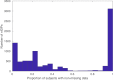




Comment in
-
The many facets of brain aging.Elife. 2020 Apr 16;9:e56640. doi: 10.7554/eLife.56640. Elife. 2020. PMID: 32297862 Free PMC article.
References
-
- Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G, Sotiropoulos SN, Jbabdi S, Hernandez-Fernandez M, Vallee E, Vidaurre D, Webster M, McCarthy P, Rorden C, Daducci A, Alexander DC, Zhang H, Dragonu I, Matthews PM, Miller KL, Smith SM. Image processing and quality control for the first 10,000 brain imaging datasets from UK biobank. NeuroImage. 2018;166:400–424. doi: 10.1016/j.neuroimage.2017.10.034. - DOI - PMC - PubMed
-
- Battle A, Brown CD, Engelhardt BE, Montgomery SB, GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts: Laboratory, Data Analysis &Coordinating Center (LDACC): NIH program management: Biospecimen collection: Pathology: eQTL manuscript working group: Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. - DOI - PMC - PubMed
-
- Brown TT, Kuperman JM, Chung Y, Erhart M, McCabe C, Hagler DJ, Venkatraman VK, Akshoomoff N, Amaral DG, Bloss CS, Casey BJ, Chang L, Ernst TM, Frazier JA, Gruen JR, Kaufmann WE, Kenet T, Kennedy DN, Murray SS, Sowell ER, Jernigan TL, Dale AM. Neuroanatomical assessment of biological maturity. Current Biology. 2012;22:1693–1698. doi: 10.1016/j.cub.2012.07.002. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

