Regulatory Mechanism of MicroRNA Expression in Cancer
- PMID: 32138313
- PMCID: PMC7084905
- DOI: 10.3390/ijms21051723
Regulatory Mechanism of MicroRNA Expression in Cancer
Abstract
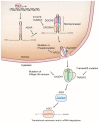
Altered gene expression is the primary molecular mechanism responsible for the pathological processes of human diseases, including cancer. MicroRNAs (miRNAs) are virtually involved at the post-transcriptional level and bind to 3' UTR of their target messenger RNA (mRNA) to suppress expression. Dysfunction of miRNAs disturbs expression of oncogenic or tumor-suppressive target genes, which is implicated in cancer pathogenesis. As such, a large number of miRNAs have been found to be downregulated or upregulated in human cancers and to function as oncomiRs or oncosuppressor miRs. Notably, the molecular mechanism underlying the dysregulation of miRNA expression in cancer has been recently uncovered. The genetic deletion or amplification and epigenetic methylation of miRNA genomic loci and the transcription factor-mediated regulation of primary miRNA often alter the landscape of miRNA expression in cancer. Dysregulation of the multiple processing steps in mature miRNA biogenesis can also cause alterations in miRNA expression in cancer. Detailed knowledge of the regulatory mechanism of miRNAs in cancer is essential for understanding its physiological role and the implications of cancer-associated dysfunction and dysregulation. In this review, we elucidate how miRNA expression is deregulated in cancer, paying particular attention to the cancer-associated transcriptional and post-transcriptional factors that execute miRNA programs.
Keywords: cancer; epigenetic modification; genetic alterations; microRNA; microRNA biogenesis; post-transcriptional regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

