Reconstructing the evolutionary history of multiple myeloma
- PMID: 32139011
- PMCID: PMC7389821
- DOI: 10.1016/j.beha.2020.101145
Reconstructing the evolutionary history of multiple myeloma
Abstract
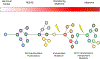
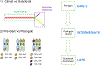
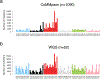
Multiple myeloma is the second most common lymphoproliferative disorder, characterized by aberrant expansion of monoclonal plasma cells. In the last years, thanks to novel next generation sequencing technologies, multiple myeloma has emerged as one of the most complex hematological cancers, shaped over time by the activity of multiple mutational processes and by the acquisition of key driver events. In this review, we describe how whole genome sequencing is emerging as a key technology to decipher this complexity at every stage of myeloma development: precursors, diagnosis and relapsed/refractory. Defining the time windows when driver events are acquired improves our understanding of cancer etiology and paves the way for early diagnosis and ultimately prevention.
Keywords: Driver events; Multiple myeloma; Timing; Whole genome sequencing.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest No conflict of interests to declare.
Figures

References
-
- Manier S, Salem KZ, Park J, Landau DA, Getz G, Ghobrial IM. Genomic complexity of multiple myeloma and its clinical implications. Nat Rev Clin Oncol. 2017;14(2):100–13. - PubMed
-
- Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335–48. - PubMed
-
- Kyle RA, Rajkumar SV. Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma. Hematol Oncol Clin North Am. 2007;21(6):1093–113, ix. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

