Genomic and serologic characterization of enterovirus A71 brainstem encephalitis
- PMID: 32139440
- PMCID: PMC7136061
- DOI: 10.1212/NXI.0000000000000703
Genomic and serologic characterization of enterovirus A71 brainstem encephalitis
Erratum in
-
Genomic and Serologic Characterization of Enterovirus A71 Brainstem Encephalitis.Neurol Neuroimmunol Neuroinflamm. 2025 Nov;12(6):e200505. doi: 10.1212/NXI.0000000000200505. Epub 2025 Oct 6. Neurol Neuroimmunol Neuroinflamm. 2025. PMID: 41052379 Free PMC article. No abstract available.
Abstract
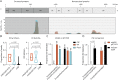
Objective: In 2016, Catalonia experienced a pediatric brainstem encephalitis outbreak caused by enterovirus A71 (EV-A71). Conventional testing identified EV in the periphery but rarely in CSF. Metagenomic next-generation sequencing (mNGS) and CSF pan-viral serology (VirScan) were deployed to enhance viral detection and characterization.
Methods: RNA was extracted from the CSF (n = 20), plasma (n = 9), stool (n = 15), and nasopharyngeal samples (n = 16) from 10 children with brainstem encephalitis and 10 children with meningitis or encephalitis. Pathogens were identified using mNGS. Available CSF from cases (n = 12) and pediatric other neurologic disease controls (n = 54) were analyzed with VirScan with a subset (n = 9 and n = 50) validated by ELISA.
Results: mNGS detected EV in all samples positive by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (n = 25). In qRT-PCR-negative samples (n = 35), mNGS found virus in 23% (n = 8, 3 CSF samples). Overall, mNGS enhanced EV detection from 42% (25/60) to 57% (33/60) (p-value = 0.013). VirScan and ELISA increased detection to 92% (11/12) compared with 46% (4/12) for CSF mNGS and qRT-PCR (p-value = 0.023). Phylogenetic analysis confirmed the EV-A71 strain clustered with a neurovirulent German EV-A71. A single amino acid substitution (S241P) in the EVA71 VP1 protein was exclusive to the CNS in one subject.
Conclusion: mNGS with VirScan significantly increased the CNS detection of EVs relative to qRT-PCR, and the latter generated an antigenic profile of the acute EV-A71 immune response. Genomic analysis confirmed the close relation of the outbreak EV-A71 and neuroinvasive German EV-A71. A S241P substitution in VP1 was found exclusively in the CSF.
Copyright © 2020 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology.
Figures


References
-
- Casas-Alba D, de Sevilla MF, Valero-Rello A, et al. Outbreak of brainstem encephalitis associated with enterovirus-A71 in Catalonia, Spain (2016): a clinical observational study in a children's reference centre in Catalonia. Clin Microbiol Infect 2017;23:874–881. - PubMed
-
- Karrasch M, Fischer E, Scholten M, et al. A severe pediatric infection with a novel enterovirus A71 strain, Thuringia, Germany. J Clin Virol 2016;84:90–95. - PubMed
