Long Non-Coding RNA Taurine Upregulated Gene 1 (TUG1) Downregulation Constrains Cell Proliferation and Invasion through Regulating Cell Division Cycle 42 (CDC42) Expression Via MiR-498 in Esophageal Squamous Cell Carcinoma Cells
- PMID: 32139664
- PMCID: PMC7077061
- DOI: 10.12659/MSM.919714
Long Non-Coding RNA Taurine Upregulated Gene 1 (TUG1) Downregulation Constrains Cell Proliferation and Invasion through Regulating Cell Division Cycle 42 (CDC42) Expression Via MiR-498 in Esophageal Squamous Cell Carcinoma Cells
Abstract
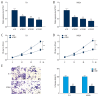
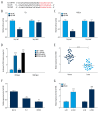
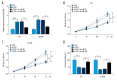
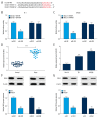
BACKGROUND Esophageal squamous cell carcinoma (ESCC) is a malignant tumor of the gastrointestinal tract. Taurine upregulated gene 1 (TUG1), a long non-coding (lnc) RNA, also known as LIN00080 or TI-227H, was connected with the tumorigenesis of various diseases. Hence, we plumed the role and molecular mechanism of TUG1 in the progression of ESCC. MATERIAL AND METHODS Expression patterns of TUG1, microRNA-498 (miR-498), and cell division cycle 42 (CDC42) mRNA were assessed using quantitative real time polymerase chain reaction (qRT-PCR). The expression level of CDC42 protein was evaluated via western blot analysis. Cell proliferation and invasion were determined with Cell Counting Kit-8 (CCK-8) assay or Transwell assay. The relationship between miR-498 and TUG1 or CDC42 was predicted by online bioinformatics database LncBase Predicted v.2 or microT-CDS and confirmed through dual-luciferase reporter system or RNA immunoprecipitation assay (RIP). RESULTS TUG1 and CDC42 were upregulated while miR-498 was strikingly decreased in ESCC tissues and cells (P<0.0001). Besides, TUG1 suppression blocked the proliferation and invasion of ESCC cells (P<0.001). Importantly, TUG1 decrease restrained CDC42 expression via binding to miR-498 in ESCC cells. Also, the suppressive impacts of TUG1 silencing on the proliferation and invasion of ESCC cells were mitigated by miR-498 reduction. Meanwhile, the repression of proliferation and invasion induced by miR-498 elevation was weakened by CDC42 overexpression. CONCLUSIONS Inhibition of TUG1 hampered cell proliferation and invasion by downregulating CDC42 via upregulating miR-498 in ESCC cells. Thus, TUG1 might be an underlying therapeutic target for ESCC.
Conflict of interest statement
None.
Figures


References
-
- Kollarova H, Machova L, Horakova D, et al. Epidemiology of esophageal cancer – an overview article. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2007;151:17–20. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. Cancer J Clin. 2015;65:5–29. - PubMed
-
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet. 2013;381:400–12. - PubMed
-
- Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–509. - PubMed
-
- Zhu ZJ, Hu Y, Zhao YF, et al. Early recurrence and death after esophagectomy in patients with esophageal squamous cell carcinoma. Ann Thorac Surg. 2011;91:1502–8. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

