Genome-wide identification and characterization of the lateral organ boundaries domain gene family in Brassica rapa var . rapa
- PMID: 32140637
- PMCID: PMC7046510
- DOI: 10.1016/j.pld.2019.11.004
Genome-wide identification and characterization of the lateral organ boundaries domain gene family in Brassica rapa var . rapa
Abstract
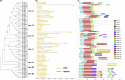
The Lateral Organ Boundaries Domain (LBD) genes encode highly conserved plant-specific LOB domain proteins which regulate growth and development in various species. However, members of the LBD gene family have yet to be identified in Brassica rapa var. rapa. In the present study, fifty-nine LBD genes were identified and distributed on 10 chromosomes. The BrrLBD proteins are predicted to encode hydrophobic polypeptides between 118 and 394 amino acids in length and with molecular weights ranging from 13.31 to 44.24 kDa; the theoretical pI for these proteins varies from 4.83 to 9.68. There were 17 paralogous gene pairs in the BrrLBD family, suggesting that the amplification of the BrrLBD gene family involved large-scale gene duplication events. Members of the BrrLBD family were divided into 7 subclades (class I a to e, class II a and b). Analysis of gene structure and conserved domains revealed that most BrrLBD genes of the same subclade had similar gene structures and protein motifs. The expression profiles of 59 BrrLBD genes were determined through Quantitative Real-time fluorescent PCR (qRT-PCR). Most BrrLBD genes in the same subclade had similar gene expression profiles. However, the expression patterns of 7 genes differed from their duplicates, indicating that although the gene function of most BrrLBD genes has been conserved, some BrrLBD genes may have undergone evolutionary change.
Keywords: Brassica rapa var. rapa; Expression profiles; LBD; LBD gene sequence analysis; Transcription factors.
© 2019 Kunming Institute of Botany, Chinese Academy of Sciences. Publishing services by Elsevier B.V. on behalf of KeAi Communications Co., Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures




References
-
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. - PubMed
-
- Cabrera J., Diaz-Manzano F.E., Sanchez M., Rosso M.N., Melillo T., Goh T., Fukaki H., Cabello S., Hofmann J., Fenoll C. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 2014;203:632–645. - PubMed
-
- Cao H., Liu C.Y., Liu C.X., Zhao Y.L., Xu R.R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet. 2016;95:515–526. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

