Fluctuation-Based Super-Resolution Traction Force Microscopy
- PMID: 32142297
- PMCID: PMC7146861
- DOI: 10.1021/acs.nanolett.9b04083
Fluctuation-Based Super-Resolution Traction Force Microscopy
Abstract
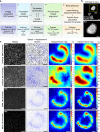
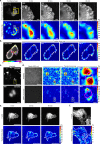
Cellular mechanics play a crucial role in tissue homeostasis and are often misregulated in disease. Traction force microscopy is one of the key methods that has enabled researchers to study fundamental aspects of mechanobiology; however, traction force microscopy is limited by poor resolution. Here, we propose a simplified protocol and imaging strategy that enhances the output of traction force microscopy by increasing i) achievable bead density and ii) the accuracy of bead tracking. Our approach relies on super-resolution microscopy, enabled by fluorescence fluctuation analysis. Our pipeline can be used on spinning-disk confocal or widefield microscopes and is compatible with available analysis software. In addition, we demonstrate that our workflow can be used to gain biologically relevant information and is suitable for fast long-term live measurement of traction forces even in light-sensitive cells. Finally, using fluctuation-based traction force microscopy, we observe that filopodia align to the force field generated by focal adhesions.
Keywords: Fluctuation-based super-resolution microscopy; SACD; SRRF; live imaging; mechanobiology; traction force microscopy.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

