OncoMX: A Knowledgebase for Exploring Cancer Biomarkers in the Context of Related Cancer and Healthy Data
- PMID: 32142370
- PMCID: PMC7101249
- DOI: 10.1200/CCI.19.00117
OncoMX: A Knowledgebase for Exploring Cancer Biomarkers in the Context of Related Cancer and Healthy Data
Abstract
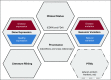
Purpose: The purpose of OncoMX1 knowledgebase development was to integrate cancer biomarker and relevant data types into a meta-portal, enabling the research of cancer biomarkers side by side with other pertinent multidimensional data types.
Methods: Cancer mutation, cancer differential expression, cancer expression specificity, healthy gene expression from human and mouse, literature mining for cancer mutation and cancer expression, and biomarker data were integrated, unified by relevant biomedical ontologies, and subjected to rule-based automated quality control before ingestion into the database.
Results: OncoMX provides integrated data encompassing more than 1,000 unique biomarker entries (939 from the Early Detection Research Network [EDRN] and 96 from the US Food and Drug Administration) mapped to 20,576 genes that have either mutation or differential expression in cancer. Sentences reporting mutation or differential expression in cancer were extracted from more than 40,000 publications, and healthy gene expression data with samples mapped to organs are available for both human genes and their mouse orthologs.
Conclusion: OncoMX has prioritized user feedback as a means of guiding development priorities. By mapping to and integrating data from several cancer genomics resources, it is hoped that OncoMX will foster a dynamic engagement between bioinformaticians and cancer biomarker researchers. This engagement should culminate in a community resource that substantially improves the ability and efficiency of exploring cancer biomarker data and related multidimensional data.
Conflict of interest statement
Marc Robinson-Rechavi
Raja Mazumder
No other potential conflicts of interest were reported.
Figures

References
-
- OncoMX https://www.oncomx.org/
-
- National Cancer Institute NCI dictionary of cancer terms. https://www.cancer.gov/publications/dictionaries/cancer-terms
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

