Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation
- PMID: 32142682
- PMCID: PMC7366517
- DOI: 10.1016/j.stem.2020.02.002
Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation
Abstract
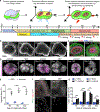
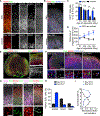
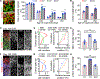
Human brain organoids provide unique platforms for modeling development and diseases by recapitulating the architecture of the embryonic brain. However, current organoid methods are limited by interior hypoxia and cell death due to insufficient surface diffusion, preventing generation of architecture resembling late developmental stages. Here, we report the sliced neocortical organoid (SNO) system, which bypasses the diffusion limit to prevent cell death over long-term cultures. This method leads to sustained neurogenesis and formation of an expanded cortical plate that establishes distinct upper and deep cortical layers for neurons and astrocytes, resembling the third trimester embryonic human neocortex. Using the SNO system, we further identify a critical role of WNT/β-catenin signaling in regulating human cortical neuron subtype fate specification, which is disrupted by a psychiatric-disorder-associated genetic mutation in patient induced pluripotent stem cell (iPSC)-derived SNOs. These results demonstrate the utility of SNOs for investigating previously inaccessible human-specific, late-stage cortical development and disease-relevant mechanisms.
Keywords: Brain organoid; DISC1; WNT; cerebral cortex; forebrain organoid; human iPSC; lamination; neurodevelopment; neuron fate specification; schizophrenia.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




References
-
- Abu-Khalil A, Fu L, Grove EA, Zecevic N, and Geschwind DH (2004). Wnt genes define distinct boundaries in the developing human brain: implications for human forebrain patterning. J. Comp. Neurol. 474, 276–288. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

