Identification and characterization of linear B cell epitopes on the nucleocapsid protein of porcine epidemic diarrhea virus using monoclonal antibodies
- PMID: 32142743
- PMCID: PMC7114562
- DOI: 10.1016/j.virusres.2020.197912
Identification and characterization of linear B cell epitopes on the nucleocapsid protein of porcine epidemic diarrhea virus using monoclonal antibodies
Abstract
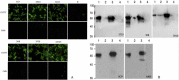
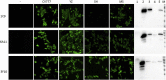
The nucleocapsid (N) protein of porcine epidemic diarrhea virus (PEDV), the most important pathogen causing severe diarrhea in piglets, is a highly conserved structural protein. In this study, 5 monoclonal antibodies (McAbs) against the PEDV N-protein were prepared and identified. Three new epitopes, 56QIRWRMRRGERI67, 318GYAQIASLAPNVAALLFGGNVA VRE342 and 398HEEAIYDDV406, were firstly identified in the viral N-protein, by using McAbs 3F10, 6A11, and 1C9. The epitope 398HEEAIYDDV406 was deleted in SH strain (isolated by our lab) and different between CV777 and YZ strain (isolated by our lab). To study the characters of this epitope, four peptides were synthesized according to the sequence of SH and CV777 and used in the study. The result showed that the 398th amino acid maybe an important amino acid of the epitope. Biological information analysis showed that the three B cell linear epitopes are highly conserved among different PEDV isolates. In addition, McAb 1C9, which attached to the epitope 398HEEAIYDDV406, showed variant reactivity with PEDV CV777, SH, YZ and MS strains. McAb 1C9 reacted with PEDV strains CV777 and YZ, but not with SH which had a deletion from 399 to 410 amino acids in N-protein (No. MK841494). Among the three McAbs, 6A11, 3F10 and 1C9, only 6A11 reacted with porcine transmissible gastroenteritis virus (TGEV) in immunofluorescence assay, therefore the other two could be used to distinguish TGEV and PEDV. These mAbs and their defined epitopes may provide useful tool for the study of the PEDV N-protein structure and function, and facilitate the development of diagnostic methods for PEDV.
Keywords: Epitopes; McAbs; Nucleocapsid protein; Porcine epidemic diarrhea virus (PEDV).
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures






Similar articles
-
Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains.J Virol. 2015 Mar;89(6):3332-42. doi: 10.1128/JVI.03196-14. Epub 2015 Jan 14. J Virol. 2015. PMID: 25589635 Free PMC article.
-
Identification and characterization of new B cell epitopes on the nucleocapsid protein of porcine epidemic diarrhea virus using monoclonal antibodies.Vet Microbiol. 2024 Nov;298:110200. doi: 10.1016/j.vetmic.2024.110200. Epub 2024 Aug 8. Vet Microbiol. 2024. PMID: 39173399
-
Identification of a novel linear B-cell epitope within the collagenase equivalent domain of porcine epidemic diarrhea virus spike glycoprotein.Virus Res. 2019 Jun;266:34-42. doi: 10.1016/j.virusres.2019.04.003. Epub 2019 Apr 6. Virus Res. 2019. PMID: 30965063
-
Identification of a conserved linear B-cell epitope in the M protein of porcine epidemic diarrhea virus.Virol J. 2012 Oct 1;9:225. doi: 10.1186/1743-422X-9-225. Virol J. 2012. PMID: 23025700 Free PMC article.
-
A Comprehensive View on the Host Factors and Viral Proteins Associated With Porcine Epidemic Diarrhea Virus Infection.Front Microbiol. 2021 Dec 7;12:762358. doi: 10.3389/fmicb.2021.762358. eCollection 2021. Front Microbiol. 2021. PMID: 34950116 Free PMC article. Review.
Cited by
-
Development of an indirect ELISA to detect PEDV specific IgA antibody based on a PEDV epidemic strain.BMC Vet Res. 2022 Aug 18;18(1):319. doi: 10.1186/s12917-022-03419-w. BMC Vet Res. 2022. PMID: 35982455 Free PMC article.
-
Correlation between the IgG/IgA Antibody Response against PEDV Structural Protein and Virus Neutralization.Microbiol Spectr. 2023 Jun 15;11(3):e0523322. doi: 10.1128/spectrum.05233-22. Epub 2023 Apr 6. Microbiol Spectr. 2023. PMID: 37022185 Free PMC article.
-
A Comprehensive View on the Protein Functions of Porcine Epidemic Diarrhea Virus.Genes (Basel). 2024 Jan 26;15(2):165. doi: 10.3390/genes15020165. Genes (Basel). 2024. PMID: 38397155 Free PMC article. Review.
-
Identification of Three Novel Linear B-Cell Epitopes in Non-Structural Protein 3 of Porcine Epidemic Diarrhea Virus Using Monoclonal Antibodies.Viruses. 2024 Mar 9;16(3):424. doi: 10.3390/v16030424. Viruses. 2024. PMID: 38543789 Free PMC article.
-
Identification of a novel B cell epitope on the nucleocapsid protein of porcine deltacoronavirus.Virus Res. 2021 Sep;302:198497. doi: 10.1016/j.virusres.2021.198497. Epub 2021 Jul 1. Virus Res. 2021. PMID: 34217778 Free PMC article.
References
-
- Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H., Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

