Reciprocal monoallelic expression of ASAR lncRNA genes controls replication timing of human chromosome 6
- PMID: 32144193
- PMCID: PMC7266157
- DOI: 10.1261/rna.073114.119
Reciprocal monoallelic expression of ASAR lncRNA genes controls replication timing of human chromosome 6
Abstract
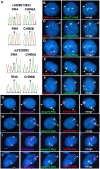
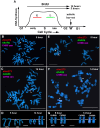
DNA replication occurs on mammalian chromosomes in a cell-type distinctive temporal order known as the replication timing program. We previously found that disruption of the noncanonical lncRNA genes ASAR6 and ASAR15 results in delayed replication timing and delayed mitotic chromosome condensation of human chromosomes 6 and 15, respectively. ASAR6 and ASAR15 display random monoallelic expression and display asynchronous replication between alleles that is coordinated with other random monoallelic genes on their respective chromosomes. Disruption of the expressed allele, but not the silent allele, of ASAR6 leads to delayed replication, activation of the previously silent alleles of linked monoallelic genes, and structural instability of human chromosome 6. In this report, we describe a second lncRNA gene (ASAR6-141) on human chromosome 6 that when disrupted results in delayed replication timing in cisASAR6-141 is subject to random monoallelic expression and asynchronous replication and is expressed from the opposite chromosome 6 homolog as ASAR6 ASAR6-141 RNA, like ASAR6 and ASAR15 RNAs, contains a high L1 content and remains associated with the chromosome territory where it is transcribed. Three classes of cis-acting elements control proper chromosome function in mammals: origins of replication, centromeres, and telomeres, which are responsible for replication, segregation, and stability of all chromosomes. Our work supports a fourth type of essential chromosomal element, the "Inactivation/Stability Center," which expresses ASAR lncRNAs responsible for proper replication timing, monoallelic expression, and structural stability of each chromosome.
Keywords: cis-acting element; noncoding RNA; replication timing.
© 2020 Heskett et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
