Evolution, expression, and substrate specificities of aldehyde oxidase enzymes in eukaryotes
- PMID: 32144208
- PMCID: PMC7170512
- DOI: 10.1074/jbc.REV119.007741
Evolution, expression, and substrate specificities of aldehyde oxidase enzymes in eukaryotes
Abstract
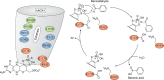
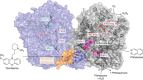
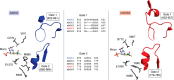
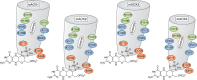
Aldehyde oxidases (AOXs) are a small group of enzymes belonging to the larger family of molybdo-flavoenzymes, along with the well-characterized xanthine oxidoreductase. The two major types of reactions that are catalyzed by AOXs are the hydroxylation of heterocycles and the oxidation of aldehydes to their corresponding carboxylic acids. Different animal species have different complements of AOX genes. The two extremes are represented in humans and rodents; whereas the human genome contains a single active gene (AOX1), those of rodents, such as mice, are endowed with four genes (Aox1-4), clustering on the same chromosome, each encoding a functionally distinct AOX enzyme. It still remains enigmatic why some species have numerous AOX enzymes, whereas others harbor only one functional enzyme. At present, little is known about the physiological relevance of AOX enzymes in humans and their additional forms in other mammals. These enzymes are expressed in the liver and play an important role in the metabolisms of drugs and other xenobiotics. In this review, we discuss the expression, tissue-specific roles, and substrate specificities of the different mammalian AOX enzymes and highlight insights into their physiological roles.
Keywords: 2Fe-2S cluster; aldehyde oxidase (AOX); drug metabolism; enzyme evolution; flavin adenine dinucleotide (FAD); flavoprotein; iron-sulfur protein; metal-containing enzyme; metalloenzyme; molybdenum; molybdenum cofactor (Moco); molybdo-flavoenzyme; mouse; oxidase; oxygen radicals; xenobiotic.
© 2020 Terao et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

