Transcriptomic and computational analysis identified LPA metabolism, KLHL14 and KCNE3 as novel regulators of Epithelial-Mesenchymal Transition
- PMID: 32144311
- PMCID: PMC7060278
- DOI: 10.1038/s41598-020-61017-y
Transcriptomic and computational analysis identified LPA metabolism, KLHL14 and KCNE3 as novel regulators of Epithelial-Mesenchymal Transition
Abstract
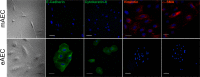
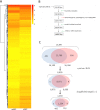
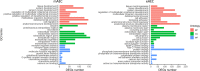
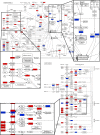
Epithelial-mesenchymal transition (EMT) is a complex biological program between physiology and pathology. Here, amniotic epithelial cells (AEC) were used as in vitro model of transiently inducible EMT in order to evaluate the transcriptional insights underlying this process. Therefore, RNA-seq was used to identify the differentially expressed genes and enrichment analyses were carried out to assess the intracellular pathways involved. As a result, molecules exclusively expressed in AEC that experienced EMT (GSTA1-1 and GSTM3) or when this process is inhibited (KLHL14 and KCNE3) were identified. Lastly, the network theory was used to obtain a computational model able to recognize putative controller genes involved in the induction and in the prevention of EMT. The results suggested an opposite role of lysophosphatidic acid (LPA) synthesis and degradation enzymes in the regulation of EMT process. In conclusion, these molecules may represent novel EMT regulators and also targets for developing new therapeutic strategies.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
High-throughput mRNA and miRNA profiling of epithelial-mesenchymal transition in MDCK cells.BMC Genomics. 2015 Nov 16;16:944. doi: 10.1186/s12864-015-2036-9. BMC Genomics. 2015. PMID: 26572553 Free PMC article.
-
Exploring the role of sphingolipid machinery during the epithelial to mesenchymal transition program using an integrative approach.Oncotarget. 2016 Apr 19;7(16):22295-323. doi: 10.18632/oncotarget.7947. Oncotarget. 2016. PMID: 26967245 Free PMC article.
-
Transcriptome of the GSH-Depleted Lens Reveals Changes in Detoxification and EMT Signaling Genes, Transport Systems, and Lipid Homeostasis.Invest Ophthalmol Vis Sci. 2017 May 1;58(5):2666-2684. doi: 10.1167/iovs.16-21398. Invest Ophthalmol Vis Sci. 2017. PMID: 28525556 Free PMC article.
-
MicroRNAs as critical regulators involved in regulating epithelial- mesenchymal transition.Curr Cancer Drug Targets. 2013 Nov;13(9):935-44. doi: 10.2174/15680096113136660099. Curr Cancer Drug Targets. 2013. PMID: 24168189 Review.
-
Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition.Cells. 2019 Sep 30;8(10):1178. doi: 10.3390/cells8101178. Cells. 2019. PMID: 31575017 Free PMC article. Review.
Cited by
-
Two-Dimensional-PAGE Coupled with nLC-MS/MS-Based Identification of Differentially Expressed Proteins and Tumorigenic Pathways in MCF7 Breast Cancer Cells Transfected for JTB Protein Silencing.Molecules. 2023 Nov 9;28(22):7501. doi: 10.3390/molecules28227501. Molecules. 2023. PMID: 38005222 Free PMC article.
-
GSTM3 Function and Polymorphism in Cancer: Emerging but Promising.Cancer Manag Res. 2020 Oct 20;12:10377-10388. doi: 10.2147/CMAR.S272467. eCollection 2020. Cancer Manag Res. 2020. PMID: 33116892 Free PMC article. Review.
-
ZEB2 haploinsufficient Mowat-Wilson syndrome induced pluripotent stem cells show disrupted GABAergic transcriptional regulation and function.Front Mol Neurosci. 2022 Oct 24;15:988993. doi: 10.3389/fnmol.2022.988993. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36353360 Free PMC article.
-
In Vitro Differentiation of Human Amniotic Epithelial Cells into Hepatocyte-like Cells.Cells. 2022 Jul 7;11(14):2138. doi: 10.3390/cells11142138. Cells. 2022. PMID: 35883581 Free PMC article.
-
Characterization of KLHL14 anti-oncogenic action in malignant mesothelioma.Heliyon. 2024 Mar 9;10(6):e27731. doi: 10.1016/j.heliyon.2024.e27731. eCollection 2024 Mar 30. Heliyon. 2024. PMID: 38509883 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

