Aryl hydrocarbon receptor-induced activation of the human IGH hs1.2 enhancer: Mutational analysis of putative regulatory binding motifs
- PMID: 32146146
- PMCID: PMC7103136
- DOI: 10.1016/j.molimm.2020.02.002
Aryl hydrocarbon receptor-induced activation of the human IGH hs1.2 enhancer: Mutational analysis of putative regulatory binding motifs
Abstract
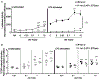
The human hs1.2 enhancer within the Ig heavy chain gene (IGH) is polymorphic and associated with a number of autoimmune diseases. The polymorphic region is characterized by tandem repeats of an ∼53-bp invariant sequence containing possible binding sites for several transcription factors. Our previous studies suggest the human hs1.2 enhancer is sensitive to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), an environmental toxicant and high affinity ligand of the aryl hydrocarbon receptor (AhR). TCDD induced hs1.2 enhancer activity in an AhR-dependent manner and the number of invariant sequences influenced the magnitude of activity. To better understand the regulation of human hs1.2 enhancer activity, the objective of the current study was to utilize mutational analysis and luciferase reporter constructs to evaluate the contribution of putative transcription factor binding sites to overall hs1.2 enhancer activity and modulation by TCDD. Basal and LPS-induced activity of the hs1.2 enhancer appeared to be most affected by mutation of sites outside of the invariant sequence or deletion of the entire invariant sequence; whereas sites influencing the effect of TCDD were dependent on the cellular activation state (i.e. unstimulated vs. LPS stimulation) and relatively independent of the putative AhR binding site within the invariant sequence. These results suggest that AhR activation affects human hs1.2 activity through an as yet undetermined non-canonical pathway. A better understanding regarding the role of the hs1.2 enhancer in human Ig expression and how AhR ligands modulate its activity may lead to insights into overall Ig regulation and mechanisms of dysfunction.
Keywords: 3′IGH regulatory region; Aryl hydrocarbon receptor; Gene regulation; Immunoglobulin; TCDD; Transcription factors.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures






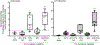

Similar articles
-
2,3,7,8-tetrachlorodibenzo-p-dioxin induces transcriptional activity of the human polymorphic hs1,2 enhancer of the 3'Igh regulatory region.J Immunol. 2012 Apr 1;188(7):3294-306. doi: 10.4049/jimmunol.1101111. Epub 2012 Feb 22. J Immunol. 2012. PMID: 22357631 Free PMC article.
-
The AhR and NF-κB/Rel Proteins Mediate the Inhibitory Effect of 2,3,7,8-Tetrachlorodibenzo-p-Dioxin on the 3' Immunoglobulin Heavy Chain Regulatory Region.Toxicol Sci. 2015 Dec;148(2):443-59. doi: 10.1093/toxsci/kfv193. Epub 2015 Sep 16. Toxicol Sci. 2015. PMID: 26377645 Free PMC article.
-
Diverse chemicals including aryl hydrocarbon receptor ligands modulate transcriptional activity of the 3'immunoglobulin heavy chain regulatory region.Toxicology. 2009 Jun 30;261(1-2):9-18. doi: 10.1016/j.tox.2009.03.015. Epub 2009 Mar 31. Toxicology. 2009. PMID: 19447539 Free PMC article.
-
Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor.Arch Biochem Biophys. 2005 Jan 15;433(2):379-86. doi: 10.1016/j.abb.2004.09.031. Arch Biochem Biophys. 2005. PMID: 15581594 Review.
-
Human response to dioxin: aryl hydrocarbon receptor (AhR) molecular structure, function, and dose-response data for enzyme induction indicate an impaired human AhR.J Toxicol Environ Health B Crit Rev. 2006 Mar-Apr;9(2):147-71. doi: 10.1080/15287390500196487. J Toxicol Environ Health B Crit Rev. 2006. PMID: 16613807 Review.
Cited by
-
Evolutive emergence and divergence of an Ig regulatory node: An environmental sensor getting cues from the aryl hydrocarbon receptor?Front Immunol. 2023 Feb 3;14:996119. doi: 10.3389/fimmu.2023.996119. eCollection 2023. Front Immunol. 2023. PMID: 36817426 Free PMC article. Review.
-
The Ah Receptor from Toxicity to Therapeutics: Report from the 5th AHR Meeting at Penn State University, USA, June 2022.Int J Mol Sci. 2023 Mar 14;24(6):5550. doi: 10.3390/ijms24065550. Int J Mol Sci. 2023. PMID: 36982624 Free PMC article.
-
Variation of the 3'RR1 HS1.2 Enhancer and Its Genomic Context.Genes (Basel). 2024 Jun 29;15(7):856. doi: 10.3390/genes15070856. Genes (Basel). 2024. PMID: 39062635 Free PMC article.
References
-
- Arnold LW, LoCascio NJ, Lutz PM, Pennell CA, Klapper D and Haughton G (1983). “Antigen-induced lymphomagenesis: identification of a murine B cell lymphoma with known antigen specificity.” J Immunol 131(4): 2064–2068. - PubMed
-
- Aupetit C, Drouet M, Pinaud E, Denizot Y, Aldigier JC, Bridoux F and Cogne M (2000). “Alleles of the alpha1 immunoglobulin gene 3′ enhancer control evolution of IgA nephropathy toward renal failure.” Kidney Int 58(3): 966–971. - PubMed
-
- Bishop GA (1992). “Signaling to a CD5+ B-cell clone through surface Ig and MHC class II molecules.” Ann N Y Acad Sci 651: 228–240. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

