Overexpression of ginseng patatin-related phospholipase pPLAIIIβ alters the polarity of cell growth and decreases lignin content in Arabidopsis
- PMID: 32148415
- PMCID: PMC7031755
- DOI: 10.1016/j.jgr.2019.01.004
Overexpression of ginseng patatin-related phospholipase pPLAIIIβ alters the polarity of cell growth and decreases lignin content in Arabidopsis
Abstract
Background: The patatin-related phospholipase AIII family (pPLAIIIs) genes alter cell elongation and cell wall composition in Arabidopsis and rice plant, suggesting diverse commercial purposes of the economically important medicinal ginseng plant. Herein, we show the functional characterization of a ginseng pPLAIII gene for the first time and discuss its potential applications.
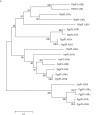
Methods: pPLAIIIs were identified from ginseng expressed sequence tag clones and further confirmed by search against ginseng database and polymerase chain reaction. A clone showing the highest homology with pPLAIIIβ was shown to be overexpressed in Arabidopsis using Agrobacterium. Quantitative polymerase chain reaction was performed to analyze ginseng pPLAIIIβ expression. Phenotypes were observed using a low-vacuum scanning electron microscope. Lignin was stained using phloroglucinol and quantified using acetyl bromide.
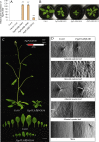
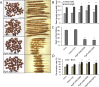
Results: The PgpPLAIIIβ transcripts were observed in all organs of 2-year-old ginseng. Overexpression of ginseng pPLAIIIβ (PgpPLAIIIβ-OE) in Arabidopsis resulted in small and stunted plants. It shortened the trichomes and decreased trichome number, indicating defects in cell polarity. Furthermore, OE lines exhibited enlarged seeds with less number per silique. The YUCCA9 gene was downregulated in the OE lines, which is reported to be associated with lignification. Accordingly, lignin was stained less in the OE lines, and the expression of two transcription factors related to lignin biosynthesis was also decreased significantly.
Conclusion: Overexpression of pPLAIIIβ retarded cell elongation in all the tested organs except seeds, which were longer and thicker than those of the controls. Shorter root length is related to auxin-responsive genes, and its stunted phenotype showed decreased lignin content.
Keywords: Auxin; Cell elongation; Lignin; Panax ginseng; Patatin-related phospholipase.
© 2019 The Korean Society of Ginseng, Published by Elsevier Korea LLC.
Figures





References
-
- Wang X. Plant phospholipases. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:211–231. - PubMed
-
- Meijer H.J., Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. - PubMed
-
- Ryu S.B. Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 2004;9:229–235. - PubMed
-
- Matos A.R., Pham-Thi A.T. Lipid deacylating enzymes in plants: old activities, new genes. Plant Physiol Biochem. 2009;47:491–503. - PubMed
-
- Scherer G.F.E., Ryu S.B., Wang X., Matos A.R., Heitz T. Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 2010;15:693–700. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

