Comparative Genomics of Actinobacillus pleuropneumoniae Serotype 8 Reveals the Importance of Prophages in the Genetic Variability of the Species
- PMID: 32149072
- PMCID: PMC7049842
- DOI: 10.1155/2020/9354204
Comparative Genomics of Actinobacillus pleuropneumoniae Serotype 8 Reveals the Importance of Prophages in the Genetic Variability of the Species
Abstract
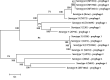
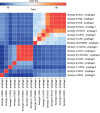
Actinobacillus pleuropneumoniae is the etiologic agent of porcine pleuropneumonia. Currently, there are 18 different serotypes; the serotype 8 is the most widely distributed in the United States, Canada, United Kingdom, and southeastern Brazil. In this study, genomes of seven A. pleuropneumoniae serotype 8 clinical isolates were compared to the other genomes of twelve serotypes. The analyses of serotype 8 genomes resulted in a set of 2352 protein-coding sequences. Of these sequences, 76.6% are present in all serotypes, 18.5% are shared with some serotypes, and 4.9% were differential. This differential portion was characterized as a series of hypothetical and regulatory protein sequences: mobile element sequence. Synteny analysis demonstrated possible events of gene recombination and acquisition by horizontal gene transfer (HGT) in this species. A total of 30 sequences related to prophages were identified in the genomes. These sequences represented 0.3 to 3.5% of the genome of the strains analyzed, and 16 of them contained complete prophages. Similarity analysis between complete prophage sequences evidenced a possible HGT with species belonging to the family Pasteurellaceae. Thus, mobile genetic elements, such as prophages, are important components of the differential portion of the A. pleuropneumoniae genome and demonstrate a central role in the evolution of the species. This study represents the first study done to understand the genome of A. pleuropneumoniae serotype 8.
Copyright © 2020 Isabelle Gonçalves de Oliveira Prado et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- OECD. Meat consumption (indicator) 2019. October 2019.
-
- Pohl S., Bertschinger H. U., Frederiksen W., Mannheim W. Transfer of Haemophilus pleuropneumoniae and the Pasteurella haemolytica-like organism causing porcine necrotic pleuropneumonia to the genus Actinobacillus (Actinobacillus pleuropneumoniae comb. nov.) on the basis of phenotypic and deoxyribonucleic acid relatedness. International Journal of Systematic and Evolutionary Microbiology. 1983;33(3):510–514. doi: 10.1099/00207713-33-3-510. - DOI
LinkOut - more resources
Full Text Sources

