Five Novel Mutations in LOXHD1 Gene Were Identified to Cause Autosomal Recessive Nonsyndromic Hearing Loss in Four Chinese Families
- PMID: 32149082
- PMCID: PMC7049443
- DOI: 10.1155/2020/1685974
Five Novel Mutations in LOXHD1 Gene Were Identified to Cause Autosomal Recessive Nonsyndromic Hearing Loss in Four Chinese Families
Erratum in
-
Corrigendum to "Five Novel Mutations in LOXHD1 Gene Were Identified to Cause Autosomal Recessive Nonsyndromic Hearing Loss in Four Chinese Families".Biomed Res Int. 2020 Sep 28;2020:9519415. doi: 10.1155/2020/9519415. eCollection 2020. Biomed Res Int. 2020. PMID: 33062705 Free PMC article.
Abstract
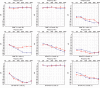
Hearing loss is one of the most common sensory disorders in newborns and is mostly caused by genetic factors. Autosomal recessive nonsyndromic hearing loss (ARNSHL) is usually characterized as a severe-to-profound congenital sensorineural hearing loss and later can cause various degrees of defect in the language and intelligent development of newborns. The mutations in LOXHD1 gene have been shown to cause DFNB77, a type of ARNSHL. To date, there are limited reports about the association between LOXHD1 gene and ARNSHL. In this study, we reported six patients from four Chinese families suffering from severe-to-profound nonsyndromic hearing loss. We performed targeted next generation sequencing in the six affected members and identified five novel pathogenic mutations in LOXHD1 including c.277G>A (p.D93N), c.611-2A>T, c.1255+3A>G, c.2329C>T (p.Q777 ∗ ), and c.5888delG (p.G1963Afs ∗ 136). These mutations were confirmed to be cosegregated with the hearing impairment in the families by Sanger sequencing and were inherited in an autosomal recessive pattern. All of the five mutations were absent in 200 control subjects. There were no symptoms of Fuchs corneal dystrophy in the probands and their blood-related relatives. We concluded that these five novel mutations could be involved in the underlying mechanism resulting in the hearing loss, and this discovery expands the genotypic spectrum of LOXHD1 mutations.
Copyright © 2020 Xiaohui Bai et al.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



References
-
- Grillet N., Schwander M., Hildebrand M. S., et al. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. The American Journal of Human Genetics. 2009;85(3):328–337. doi: 10.1016/j.ajhg.2009.07.017. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

