Comprehensive Analysis of lncRNA-Mediated ceRNA Crosstalk and Identification of Prognostic Biomarkers in Wilms' Tumor
- PMID: 32149111
- PMCID: PMC7057018
- DOI: 10.1155/2020/4951692
Comprehensive Analysis of lncRNA-Mediated ceRNA Crosstalk and Identification of Prognostic Biomarkers in Wilms' Tumor
Abstract
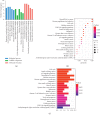
Wilms' tumor (WT) is the most common type of childhood kidney cancer, and most cases present with favorable histology and respond well to standard treatment. However, a subset of patients with WT is diagnosed with bilateral, relapsed, and high-risk tumors which remain the leading cause of cancer-related death in children. Long noncoding RNAs (lncRNAs) and their aberrant expression have currently been attracting great attention as oncogenes or tumor suppressors during tumor initiation and progression. So far, their roles and related competitive endogenous RNA (ceRNA) network remain unelucidated in nephroblastoma pathogenesis. We comprehensively integrated lncRNA, microRNA (miRNA), and messenger RNA (mRNA) expression profiles from the Therapeutically Applicable Research to Generate Effective Treatment (TARGET) database and screened out differentially expressed mRNAs (DEMs), lncRNAs (DELs), and miRNAs (DEMis) to construct a ceRNA network based on the information generated from miRcode, miRTarBase, TargetScan, and miRDB. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to analyze the functional characteristics of DEMs in the ceRNA network. The interaction between protein molecules was also analyzed by establishing a protein-protein interaction network. Finally, prognosis-related biomarkers were identified via survival analysis. Initially, 1647 DELs, 115 DEMis, and 3280 DEMs (|log FC| > 2; FDR < 0.01) were obtained using the R package. Next, we constructed a lncRNA-miRNA-mRNA network (ceRNA network), in which 176 DELs, 24 DEMis, and 141 DEMs were identified. Furthermore, 148 functional enrichment terms from GO were identified and 29 KEGG pathways were found to be significantly enriched. We also integrated patient clinical information to analyze the association between DERNAs and patient prognosis. We found that high expression of 8 DELs (LINC00473, AL445228.2, DENND5B-AS1, DLEU2, AC123595.1, AC135178.1, LINC00535, and LMO7-AS1) and 4 DEMs (CEP55, DEPDC1, PHF19, and TRIM36) correlated with poor survival in a patient with WT, whereas high expression of 2 DELs (MEG3 and RMST), 1 DEM (KIAA0922), and 1 DEMi (hsa-mir-200a) could possibly lead to better clinical outcomes. For the first time, the present study provided a novel insight into lncRNA-related ceRNA networks and identified potential prognostic biomarkers in Wilms' tumor.
Copyright © 2020 Hong Zheng et al.
Conflict of interest statement
The authors declare that they have no conflicts of interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

