Machine Learning Analysis of Image Data Based on Detailed MR Image Reports for Nasopharyngeal Carcinoma Prognosis
- PMID: 32149139
- PMCID: PMC7054759
- DOI: 10.1155/2020/8068913
Machine Learning Analysis of Image Data Based on Detailed MR Image Reports for Nasopharyngeal Carcinoma Prognosis
Abstract
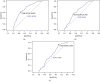
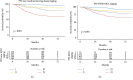
We aimed to assess the use of automatic machine learning (AutoML) algorithm based on magnetic resonance (MR) image data to assign prediction scores to patients with nasopharyngeal carcinoma (NPC). We also aimed to develop a 4-group classification system for NPC, superior to the current clinical staging system. Between January 2010 and January 2013, 792 patients with recent diagnosis of NPC, who had MR image data, were enrolled in the study. The AutoML algorithm was used and all statistical analyses were based on the 10-fold test. Primary endpoints included the probabilities of overall survival (OS), distant metastasis-free survival (DMFS), and local-region relapse-free survival (LRFS), and their sum was recorded as the final voting score, representative of progression-free survival (PFS) for each patient. The area under the receiver operating characteristic (ROC) curve generated from the MR image data-based model compared with the tumor, node, and metastasis (TNM) system-based model was 0.796 (P=0.008) for OS, 0.752 (P=0.053) for DMFS, and 0.721 (P=0.025) for LRFS. The Kaplan-Meier (KM) test values for II/I, III/II, IV/III groups in our new machine learning-based scoring system were 0.011, 0.010, and <0.001, respectively, whereas those for II/I, III/II, IV/III groups in the TNM/American Joint Committee on Cancer (AJCC) system were 0.118, 0.121, and <0.001, respectively. Significant differences were observed in the new machine learning-based scoring system analysis of each curve (P < 0.05), whereas the P values of curves obtained from the TNM/AJCC system, between II/I and III/II, were 0.118 and 0.121, respectively, without a significant difference. In conclusion, the AutoML algorithm demonstrated better prognostic performance than the TNM/AJCC system for NPC. The algorithm showed a good potential for clinical application and may aid in improving counseling and facilitate the personalized management of patients with NPC. The clinical application of our new scoring and staging system may significantly improve precision medicine.
Copyright © 2020 Chunyan Cui et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures


Similar articles
-
Radiomics on multi-modalities MR sequences can subtype patients with non-metastatic nasopharyngeal carcinoma (NPC) into distinct survival subgroups.Eur Radiol. 2019 Oct;29(10):5590-5599. doi: 10.1007/s00330-019-06075-1. Epub 2019 Mar 14. Eur Radiol. 2019. PMID: 30874880
-
Predictive value of pretreatment MRI texture analysis in patients with primary nasopharyngeal carcinoma.Eur Radiol. 2019 Aug;29(8):4105-4113. doi: 10.1007/s00330-018-5961-6. Epub 2019 Jan 7. Eur Radiol. 2019. PMID: 30617473 Free PMC article.
-
Combination of diffusion-weighted imaging and arterial spin labeling at 3.0 T for the clinical staging of nasopharyngeal carcinoma.Clin Imaging. 2020 Oct;66:127-132. doi: 10.1016/j.clinimag.2020.05.007. Epub 2020 May 15. Clin Imaging. 2020. PMID: 32480267
-
Evolution of the Chinese staging system for nasopharyngeal carcinoma.Chin Clin Oncol. 2016 Apr;5(2):19. doi: 10.21037/cco.2016.03.04. Chin Clin Oncol. 2016. PMID: 27121879 Review.
-
MRI-based radiomics models can improve prognosis prediction for nasopharyngeal carcinoma with neoadjuvant chemotherapy.Magn Reson Imaging. 2022 May;88:108-115. doi: 10.1016/j.mri.2022.02.005. Epub 2022 Feb 15. Magn Reson Imaging. 2022. PMID: 35181470
Cited by
-
Application of Artificial Intelligence to the Diagnosis and Therapy of Nasopharyngeal Carcinoma.J Clin Med. 2023 Apr 24;12(9):3077. doi: 10.3390/jcm12093077. J Clin Med. 2023. PMID: 37176518 Free PMC article. Review.
-
A Comprehensive Review on Radiomics and Deep Learning for Nasopharyngeal Carcinoma Imaging.Diagnostics (Basel). 2021 Aug 24;11(9):1523. doi: 10.3390/diagnostics11091523. Diagnostics (Basel). 2021. PMID: 34573865 Free PMC article. Review.
-
A coordinate registration-based structured magnetic resonance imaging reporting method for nasopharyngeal carcinoma: a preliminary study.Quant Imaging Med Surg. 2025 Mar 3;15(3):2444-2456. doi: 10.21037/qims-24-1127. Epub 2025 Feb 26. Quant Imaging Med Surg. 2025. PMID: 40160624 Free PMC article.
-
Application of Artificial Intelligence for Nasopharyngeal Carcinoma Management - A Systematic Review.Cancer Manag Res. 2022 Jan 26;14:339-366. doi: 10.2147/CMAR.S341583. eCollection 2022. Cancer Manag Res. 2022. PMID: 35115832 Free PMC article. Review.
-
Predictive function of tumor burden-incorporated machine-learning algorithms for overall survival and their value in guiding management decisions in patients with locally advanced nasopharyngeal carcinoma.J Natl Cancer Cent. 2023 Oct 12;3(4):295-305. doi: 10.1016/j.jncc.2023.10.002. eCollection 2023 Dec. J Natl Cancer Cent. 2023. PMID: 39036668 Free PMC article.
References
-
- Hsiung C.-Y., Yorke E. D., Chui C.-S., et al. Intensity-modulated radiotherapy versus conventional three-dimensional conformal radiotherapy for boost or salvage treatment of nasopharyngeal carcinoma. International Journal of Radiation Oncology ∗ Biology ∗ Physics. 2002;53(3):638–647. doi: 10.1016/s0360-3016(02)02760-8. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

