Current knowledge and recent advances in understanding metabolism of the model cyanobacterium Synechocystis sp. PCC 6803
- PMID: 32149336
- PMCID: PMC7133116
- DOI: 10.1042/BSR20193325
Current knowledge and recent advances in understanding metabolism of the model cyanobacterium Synechocystis sp. PCC 6803
Abstract
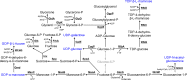
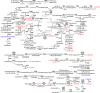
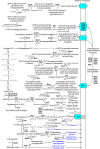
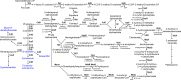
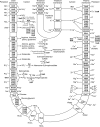
Cyanobacteria are key organisms in the global ecosystem, useful models for studying metabolic and physiological processes conserved in photosynthetic organisms, and potential renewable platforms for production of chemicals. Characterizing cyanobacterial metabolism and physiology is key to understanding their role in the environment and unlocking their potential for biotechnology applications. Many aspects of cyanobacterial biology differ from heterotrophic bacteria. For example, most cyanobacteria incorporate a series of internal thylakoid membranes where both oxygenic photosynthesis and respiration occur, while CO2 fixation takes place in specialized compartments termed carboxysomes. In this review, we provide a comprehensive summary of our knowledge on cyanobacterial physiology and the pathways in Synechocystis sp. PCC 6803 (Synechocystis) involved in biosynthesis of sugar-based metabolites, amino acids, nucleotides, lipids, cofactors, vitamins, isoprenoids, pigments and cell wall components, in addition to the proteins involved in metabolite transport. While some pathways are conserved between model cyanobacteria, such as Synechocystis, and model heterotrophic bacteria like Escherichia coli, many enzymes and/or pathways involved in the biosynthesis of key metabolites in cyanobacteria have not been completely characterized. These include pathways required for biosynthesis of chorismate and membrane lipids, nucleotides, several amino acids, vitamins and cofactors, and isoprenoids such as plastoquinone, carotenoids, and tocopherols. Moreover, our understanding of photorespiration, lipopolysaccharide assembly and transport, and degradation of lipids, sucrose, most vitamins and amino acids, and haem, is incomplete. We discuss tools that may aid our understanding of cyanobacterial metabolism, notably CyanoSource, a barcoded library of targeted Synechocystis mutants, which will significantly accelerate characterization of individual proteins.
Keywords: Synechocystis; comparative genomics; cyanobacteria; degradation; metabolism.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures





References
-
- Zwirglmaier K. et al. (2008) Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161 - PubMed
-
- Galloway J.N. et al. (2004) Nitrogen cycles: past, present, and future. Biogeochemistry 70, 153–226 10.1007/s10533-004-0370-0 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

