Metagenomics for taxonomy profiling: tools and approaches
- PMID: 32149573
- PMCID: PMC7161568
- DOI: 10.1080/21655979.2020.1736238
Metagenomics for taxonomy profiling: tools and approaches
Retraction in
-
Statement of Retraction: Metagenomics for taxonomy profiling: tools and approaches.Bioengineered. 2022 Jun;13(6):14750. doi: 10.1080/21655979.2022.2126093. Bioengineered. 2022. PMID: 36154550 Free PMC article. No abstract available.
Abstract
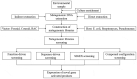
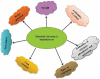
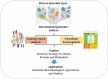
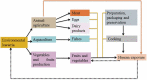
The study of metagenomics is an emerging field that identifies the total genetic materials in an organism along with the set of all genetic materials like deoxyribonucleic acid and ribose nucleic acid, which play a key role with the maintenance of cellular functions. The best part of this technology is that it gives more flexibility to environmental microbiologists to instantly pioneer the immense genetic variability of microbial communities. However, it is intensively complex to identify the suitable sequencing measures of any specific gene that can exclusively indicate the involvement of microbial metagenomes and be able to advance valuable results about these communities. This review provides an overview of the metagenomic advancement that has been advantageous for aggregation of more knowledge about specific genes, microbial communities and its metabolic pathways. More specific drawbacks of metagenomes technology mainly depend on sequence-based analysis. Therefore, this 'targeted based metagenomics' approach will give comprehensive knowledge about the ecological, evolutionary and functional sequence of significantly important genes that naturally exist in living beings either human, animal and microorganisms from distinctive ecosystems.
Keywords: deoxyribonucleic acid; genomics; metagenomics; microbial metagenomes; ribose nucleic acid.
Figures

References
-
- Handelsman J, Rondon MR, Brady SF, et al. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:245–249. - PubMed
-
- Allen EE, Banfield JF.. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–498. - PubMed
-
- Cowan DA, Arslanoglu A, Burton SG. Metagenomics, gene discovery, and the ideal biocatalyst. Biochem Soc Trans. 2004;32:298–302. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
