Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis
- PMID: 32149598
- PMCID: PMC7276701
- DOI: 10.1099/mgen.0.000347
Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis
Abstract
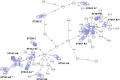
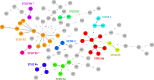
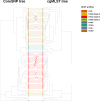
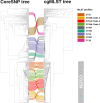
In this work we compared the most frequently used Klebsiella pneumoniae typing methods: PFGE, cgMLST and coreSNP. We evaluated the discriminatory power of the three methods to confirm or exclude nosocomial transmission on K. pneumoniae strains isolated from January to December 2017, in the framework of the routine surveillance for multidrug-resistant organisms at the San Raffaele Hospital, in Milan. We compared the results of the different methods to the results of epidemiological investigation. Our results showed that cgMLST and coreSNP are more discriminant than PFGE, and that both approaches are suitable for transmission analyses. cgMLST appeared to be inferior to coreSNP in the K. pneumoniae CG258 phylogenetic reconstruction. Indeed, we found that the phylogenetic reconstruction based on cgMLST genes wrongly clustered ST258 clade1 and clade2 strains, conversely properly assigned by coreSNP approach. In conclusion, this study provides evidences supporting the reliability of both cgMLST and coreSNP for hospital surveillance programs and highlights the limits of cgMLST scheme genes for phylogenetic reconstructions.
Keywords: CG258; K. pneumoniae; cgMLST; cluster.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
References
-
- Giani T, Antonelli A, Caltagirone M, Mauri C, Nicchi J, et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: KPC-carbapenemase spreading among outpatients. Euro Surveill. 2017;22:pii: 30583. doi: 10.2807/1560-7917.ES.2017.22.31.30583. - DOI - PMC - PubMed

