The single-cell eQTLGen consortium
- PMID: 32149610
- PMCID: PMC7077978
- DOI: 10.7554/eLife.52155
The single-cell eQTLGen consortium
Abstract
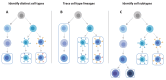
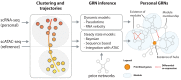
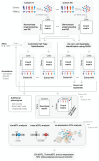
In recent years, functional genomics approaches combining genetic information with bulk RNA-sequencing data have identified the downstream expression effects of disease-associated genetic risk factors through so-called expression quantitative trait locus (eQTL) analysis. Single-cell RNA-sequencing creates enormous opportunities for mapping eQTLs across different cell types and in dynamic processes, many of which are obscured when using bulk methods. Rapid increase in throughput and reduction in cost per cell now allow this technology to be applied to large-scale population genetics studies. To fully leverage these emerging data resources, we have founded the single-cell eQTLGen consortium (sc-eQTLGen), aimed at pinpointing the cellular contexts in which disease-causing genetic variants affect gene expression. Here, we outline the goals, approach and potential utility of the sc-eQTLGen consortium. We also provide a set of study design considerations for future single-cell eQTL studies.
Keywords: PBMC; eQTL; gene regulatory network; genetics; genomics; human; science forum; single-cell.
© 2020, van der Wijst et al.
Conflict of interest statement
Mv, Dd, HG, GT, CH, MB, OS, MN, YI, Pv, CY, JP, FT, AM, MH, LF No competing interests declared
Figures


References
-
- Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, van den Oord J, Atak ZK, Wouters J, Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nature Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. - DOI - PMC - PubMed
-
- Alquicira-Hernández J, Sathe A, Ji HP, Nguyen Q, Powell JE. scPred: cell type prediction at single-cell resolution. bioRxiv. 2018 doi: 10.1101/369538. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

