Transcriptomic Analysis of the Developmental Similarities and Differences Between the Native Retina and Retinal Organoids
- PMID: 32150248
- PMCID: PMC7401421
- DOI: 10.1167/iovs.61.3.6
Transcriptomic Analysis of the Developmental Similarities and Differences Between the Native Retina and Retinal Organoids
Abstract
Purpose: We performed a bioinformatic transcriptome analysis to determine the alteration of gene expression between the native retina and retinal organoids in both mice and humans.
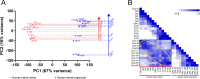
Methods: The datasets of mouse native retina (GSE101986), mouse retinal organoids (GSE102794), human native retina (GSE104827), and human retinal organoids (GSE119320) were obtained from Gene Expression Omnibus. After normalization, a principal component analysis was performed to categorize the samples. The genes were clustered to classify them. A functional analysis was performed using the bioinformatics tool Gene ontology enrichment to analyze the biological processes of selected genes and cellular components.
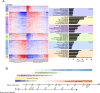
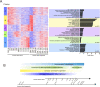
Results: The development of retinal organoids is slower than that in the native retina. In the early stage, cell proliferation predominates. Subsequently, neural differentiation is dominant. In the later stage, the dominant differentiated cells are photoreceptors. Additionally, the fatty acid metabolic process and mitochondria-related genes are upregulated over time, and the glycogen catabolic process and activin receptors are gradually downregulated in human retinal organoids. However, these trends are opposite in mouse retinal organoids. There are two peaks in mitochondria-related genes, one in the early development period and another during the photoreceptor development period. It takes about five times longer for human retinal development to achieve similar levels of mouse retinal development.
Conclusions: Our study reveals the similarities and differences in the developmental features of retinal organoids as well as the corresponding relationship between mouse and human retinal development.
Conflict of interest statement
Disclosure:
Figures





References
-
- Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012; 35: 565–573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

