The core spliceosomal factor U2AF1 controls cell-fate determination via the modulation of transcriptional networks
- PMID: 32150510
- PMCID: PMC7549707
- DOI: 10.1080/15476286.2020.1733800
The core spliceosomal factor U2AF1 controls cell-fate determination via the modulation of transcriptional networks
Abstract
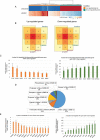
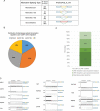
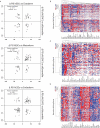
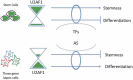
Alternative splicing (AS) plays a central role during cell-fate determination. However, how the core spliceosomal factors (CSFs) are involved in this process is poorly understood. Here, we report the down-regulation of the U2AF1 CSF during stem cell differentiation. To investigate its function in stemness and differentiation, we downregulated U2AF1 in human induced pluripotent stem cells (hiPSCs), using an inducible-shRNA system, to the level found in differentiated ectodermal, mesodermal and endodermal cells. RNA sequencing and computational analysis reveal that U2AF1 down-regulation modulates the expression of development-regulating genes and regulates transcriptional networks involved in cell-fate determination. Furthermore, U2AF1 down-regulation induces a switch in the AS of transcription factors (TFs) required to establish specific cell lineages, and favours the splicing of a differentiated cell-specific isoform of DNMT3B. Our results showed that the differential expression of the core spliceosomal factor U2AF1, between stem cells and the precursors of the three germ layers regulates a cell-type-specific alternative splicing programme and a transcriptional network involved in cell-fate determination via the modulation of gene expression and alternative splicing of transcription regulators.
Keywords: Cell-fate determination; RNA; Spliceosome; Splicing; Stem cells; Transcription.
Conflict of interest statement
The authors declare that they have no competing interests or conflict of interest, financial or otherwise.
Figures


Similar articles
-
Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome.Nat Commun. 2017 Jan 9;8:14060. doi: 10.1038/ncomms14060. Nat Commun. 2017. PMID: 28067246 Free PMC article.
-
Revealing the role of U2AF1 in splicing regulation and chimeric RNA dynamics.Sci Rep. 2025 May 9;15(1):16235. doi: 10.1038/s41598-025-99865-1. Sci Rep. 2025. PMID: 40346396 Free PMC article.
-
Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1.Proc Natl Acad Sci U S A. 2018 Oct 30;115(44):E10437-E10446. doi: 10.1073/pnas.1812669115. Epub 2018 Oct 15. Proc Natl Acad Sci U S A. 2018. PMID: 30322915 Free PMC article.
-
Splicing factor gene mutations in the myelodysplastic syndromes: impact on disease phenotype and therapeutic applications.Adv Biol Regul. 2017 Jan;63:59-70. doi: 10.1016/j.jbior.2016.08.001. Epub 2016 Aug 21. Adv Biol Regul. 2017. PMID: 27639445 Review.
-
Evolution of the Early Spliceosomal Complex-From Constitutive to Regulated Splicing.Int J Mol Sci. 2021 Nov 18;22(22):12444. doi: 10.3390/ijms222212444. Int J Mol Sci. 2021. PMID: 34830325 Free PMC article. Review.
Cited by
-
Osteoblast-Derived Mitochondria Formulated with Cationic Liposome Guide Mesenchymal Stem Cells into Osteogenic Differentiation.Adv Sci (Weinh). 2025 Mar;12(12):e2412621. doi: 10.1002/advs.202412621. Epub 2025 Jan 31. Adv Sci (Weinh). 2025. PMID: 39887937 Free PMC article.
-
Mutant U2AF1-induced differential alternative splicing causes an oxidative stress in bone marrow stromal cells.Exp Biol Med (Maywood). 2021 Aug;246(15):1750-1759. doi: 10.1177/15353702211010130. Epub 2021 May 25. Exp Biol Med (Maywood). 2021. PMID: 34034558 Free PMC article.
-
Heat Shock Alters the Proteomic Profile of Equine Mesenchymal Stem Cells.Int J Mol Sci. 2022 Jun 29;23(13):7233. doi: 10.3390/ijms23137233. Int J Mol Sci. 2022. PMID: 35806237 Free PMC article.
-
Alternative splicing in mesenchymal stem cell differentiation.Stem Cells. 2020 Oct 1;38(10):1229-1240. doi: 10.1002/stem.3248. Epub 2020 Jul 15. Stem Cells. 2020. PMID: 32627865 Free PMC article. Review.
-
The emerging significance of splicing in vertebrate development.Development. 2022 Oct 1;149(19):dev200373. doi: 10.1242/dev.200373. Epub 2022 Sep 30. Development. 2022. PMID: 36178052 Free PMC article. Review.
References
-
- Irimia M, Blencowe BJ.. Alternative splicing: decoding an expansive regulatory layer. Curr Opin Cell Biol. 2012;24:323–332. - PubMed
-
- Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. - PubMed
-
- Gabut M, Samavarchi-Tehrani P, Wang X, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. - PubMed
-
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
