Repurposing the Streptococcus mutans CRISPR-Cas9 System to Understand Essential Gene Function
- PMID: 32150575
- PMCID: PMC7082069
- DOI: 10.1371/journal.ppat.1008344
Repurposing the Streptococcus mutans CRISPR-Cas9 System to Understand Essential Gene Function
Abstract
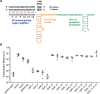
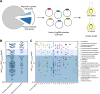
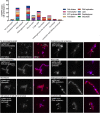
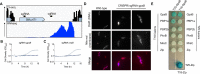
A recent genome-wide screen identified ~300 essential or growth-supporting genes in the dental caries pathogen Streptococcus mutans. To be able to study these genes, we built a CRISPR interference tool around the Cas9 nuclease (Cas9Smu) encoded in the S. mutans UA159 genome. Using a xylose-inducible dead Cas9Smu with a constitutively active single-guide RNA (sgRNA), we observed titratable repression of GFP fluorescence that compared favorably to that of Streptococcus pyogenes dCas9 (Cas9Spy). We then investigated sgRNA specificity and proto-spacer adjacent motif (PAM) requirements. Interference by sgRNAs did not occur with double or triple base-pair mutations, or if single base-pair mutations were in the 3' end of the sgRNA. Bioinformatic analysis of >450 S. mutans genomes allied with in vivo assays revealed a similar PAM recognition sequence as Cas9Spy. Next, we created a comprehensive library of sgRNA plasmids that were directed at essential and growth-supporting genes. We discovered growth defects for 77% of the CRISPRi strains expressing sgRNAs. Phenotypes of CRISPRi strains, across several biological pathways, were assessed using fluorescence microscopy. A variety of cell structure anomalies were observed, including segregational instability of the chromosome, enlarged cells, and ovococci-to-rod shape transitions. CRISPRi was also employed to observe how silencing of cell wall glycopolysaccharide biosynthesis (rhamnose-glucose polysaccharide, RGP) affected both cell division and pathogenesis in a wax worm model. The CRISPRi tool and sgRNA library are valuable resources for characterizing essential genes in S. mutans, some of which could prove to be promising therapeutic targets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

