A Phylogenetic Analysis of Hepatitis C Virus Transmission, Relapse, and Reinfection Among People Who Inject Drugs Receiving Opioid Agonist Therapy
- PMID: 32150621
- PMCID: PMC7336560
- DOI: 10.1093/infdis/jiaa100
A Phylogenetic Analysis of Hepatitis C Virus Transmission, Relapse, and Reinfection Among People Who Inject Drugs Receiving Opioid Agonist Therapy
Abstract
Background: Understanding hepatitis C virus (HCV) transmission among people who inject drugs (PWID) is essential for HCV elimination. We aimed to differentiate reinfections from treatment failures and to identify transmission linkages and associated factors in a cohort of PWID receiving opioid agonist therapy (OAT).
Methods: We analyzed baseline and follow-up specimens from 150 PWID from 3 OAT clinics in the Bronx, New York. Next-generation sequencing data from the hypervariable region 1 of HCV were analyzed using Global Hepatitis Outbreak and Surveillance Technology.
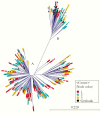
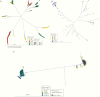
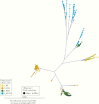
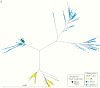
Results: There were 3 transmission linkages between study participants. Sustained virologic response (SVR) was not achieved in 9 participants: 7 had follow-up specimens with similar sequences to baseline, and 2 died. In 4 additional participants, SVR was achieved but the participants were viremic at later follow-up: 2 were reinfected with different strains, 1 had a late treatment failure, and 1 was transiently viremic 17 months after treatment. All transmission linkages were from the same OAT clinic and involved spousal or common-law partnerships.
Conclusion: This study highlights the use of next-generation sequencing as an important tool for identifying viral transmission and to help distinguish relapse and reinfection among PWID. Results reinforce the need for harm reduction interventions among couples and those who report ongoing risk factors after SVR.
Keywords: HCV; IDU; OAT; PWID; hepatitis C; next generation sequencing; phylogenetic; transmission network.
© The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures



Similar articles
-
Low Hepatitis C Reinfection Following Direct-acting Antiviral Therapy Among People Who Inject Drugs on Opioid Agonist Therapy.Clin Infect Dis. 2020 Jun 10;70(12):2695-2702. doi: 10.1093/cid/ciz693. Clin Infect Dis. 2020. PMID: 31346609 Free PMC article. Clinical Trial.
-
Hepatitis C virus reinfection after successful treatment with direct-acting antiviral therapy in a large population-based cohort.J Hepatol. 2018 Nov;69(5):1007-1014. doi: 10.1016/j.jhep.2018.07.025. Epub 2018 Aug 22. J Hepatol. 2018. PMID: 30142429
-
Hepatitis C reinfection after successful antiviral treatment among people who inject drugs: A meta-analysis.J Hepatol. 2020 Apr;72(4):643-657. doi: 10.1016/j.jhep.2019.11.012. Epub 2019 Nov 27. J Hepatol. 2020. PMID: 31785345
-
Changes in Health-related Quality of Life for Hepatitis C Virus-Infected People Who Inject Drugs While on Opioid Agonist Treatment Following Sustained Virologic Response.Clin Infect Dis. 2022 May 3;74(9):1586-1593. doi: 10.1093/cid/ciab669. Clin Infect Dis. 2022. PMID: 34331539 Free PMC article.
-
Hepatitis C Reinfection in People Who Inject Drugs in Resource-Limited Countries: A Systematic Review and Analysis.Int J Environ Res Public Health. 2020 Jul 9;17(14):4951. doi: 10.3390/ijerph17144951. Int J Environ Res Public Health. 2020. PMID: 32659974 Free PMC article.
Cited by
-
Hepatitis C Resistance-Associated Substitutions Among People Who Inject Drugs Treated With Direct-Acting Antiviral-Containing Regimens.Open Forum Infect Dis. 2021 Sep 30;8(10):ofab474. doi: 10.1093/ofid/ofab474. eCollection 2021 Oct. Open Forum Infect Dis. 2021. PMID: 34692891 Free PMC article.
-
HCV prevalence and phylogenetic characteristics in a cross-sectional, community study of young people who inject drugs in New York City: Opportunity for and threats to HCV elimination.Health Sci Rep. 2024 Jul 1;7(7):e2211. doi: 10.1002/hsr2.2211. eCollection 2024 Jul. Health Sci Rep. 2024. PMID: 38957862 Free PMC article.
-
An Increase in the Prevalence of Clinically Relevant Resistance-Associated Substitutions in Four Direct-Acting Antiviral Regimens: A Study Using GenBank HCV Sequences.Pathogens. 2024 Aug 9;13(8):674. doi: 10.3390/pathogens13080674. Pathogens. 2024. PMID: 39204274 Free PMC article.
-
Genomic surveillance uncovers regional variation in HCV transmission networks among people who use drugs in rural U.S. communities.Res Sq [Preprint]. 2025 Jun 10:rs.3.rs-6810633. doi: 10.21203/rs.3.rs-6810633/v1. Res Sq. 2025. PMID: 40585240 Free PMC article. Preprint.
-
Epidemiological data analysis of viral quasispecies in the next-generation sequencing era.Brief Bioinform. 2021 Jan 18;22(1):96-108. doi: 10.1093/bib/bbaa101. Brief Bioinform. 2021. PMID: 32568371 Free PMC article. Review.
References
-
- Verna EC, Brown RS Jr. Hepatitis C virus and liver transplantation. Clin Liver Dis 2006; 10:919–40. - PubMed
-
- US Department of Health and Human Services. Combating the silent epidemic of viral hepatitis: action plan for the prevention, care & treatment of viral hepatitis. 2011. https://www.hhs.gov/sites/default/files/action-plan-viral-hepatitis-2011.... Accessed 5 January 2020.
-
- Ly KN, Xing J, Klevens RM, Jiles RB, Ward JW, Holmberg SD. The increasing burden of mortality from viral hepatitis in the United States between 1999 and 2007. Ann Intern Med 2012; 156:271–8. - PubMed
-
- Rein DB, Wittenborn JS, Weinbaum CM, Sabin M, Smith BD, Lesesne SB. Forecasting the morbidity and mortality associated with prevalent cases of pre-cirrhotic chronic hepatitis C in the United States. Dig Liver Dis 2011; 43:66–72. - PubMed
-
- Davis GL, Alter MJ, El-Serag H, Poynard T, Jennings LW. Aging of hepatitis C virus (HCV)-infected persons in the United States: a multiple cohort model of HCV prevalence and disease progression. Gastroenterology 2010; 138:513–21, 521.e1–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

