Comparative Root Transcriptomics Provide Insights into Drought Adaptation Strategies in Chickpea (Cicer arietinum L.)
- PMID: 32150870
- PMCID: PMC7084756
- DOI: 10.3390/ijms21051781
Comparative Root Transcriptomics Provide Insights into Drought Adaptation Strategies in Chickpea (Cicer arietinum L.)
Abstract
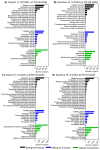
Drought adversely affects crop production across the globe. The root system immensely contributes to water management and the adaptability of plants to drought stress. In this study, drought-induced phenotypic and transcriptomic responses of two contrasting chickpea (Cicer arietinum L.) genotypes were compared at the vegetative, reproductive transition, and reproductive stages. At the vegetative stage, drought-tolerant genotype maintained higher root biomass, length, and surface area under drought stress as compared to sensitive genotype. However, at the reproductive stage, root length and surface area of tolerant genotype was lower but displayed higher root diameter than sensitive genotype. The shoot biomass of tolerant genotype was overall higher than the sensitive genotype under drought stress. RNA-seq analysis identified genotype- and developmental-stage specific differentially expressed genes (DEGs) in response to drought stress. At the vegetative stage, a total of 2161 and 1873 DEGs, and at reproductive stage 4109 and 3772 DEGs, were identified in the tolerant and sensitive genotypes, respectively. Gene ontology (GO) analysis revealed enrichment of biological categories related to cellular process, metabolic process, response to stimulus, response to abiotic stress, and response to hormones. Interestingly, the expression of stress-responsive transcription factors, kinases, ROS signaling and scavenging, transporters, root nodulation, and oxylipin biosynthesis genes were robustly upregulated in the tolerant genotype, possibly contributing to drought adaptation. Furthermore, activation/repression of hormone signaling and biosynthesis genes was observed. Overall, this study sheds new insights on drought tolerance mechanisms operating in roots with broader implications for chickpea improvement.
Keywords: abiotic stress; chickpea; drought; gene expression; hormone; root; signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Farooq M., Wahid A., Kobayashi N., Fujita D., Basra S. Sustainable Agriculture. Springer; Berlin/Heidelberg, Germany: 2009. Plant drought stress: Effects, mechanisms and management; pp. 153–188.
-
- Leport L., Turner N.C., Davies S.L., Siddique K.H.M. Variation in pod production and abortion among chickpea cultivars under terminal drought. Eur. J. Agron. 2006;24:236–246. doi: 10.1016/j.eja.2005.08.005. - DOI
-
- Fang X., Turner N.C., Yan G., Li F., Siddique K.H.M. Flower numbers, pod production, pollen viability, and pistil function are reduced and flower and pod abortion increased in chickpea (Cicer arietinum L.) under terminal drought. J. Exp. Bot. 2009;61:335–345. doi: 10.1093/jxb/erp307. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

