DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types
- PMID: 32150897
- PMCID: PMC7140532
- DOI: 10.3390/cells9030624
DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types
Abstract
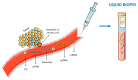
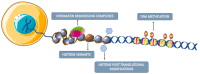
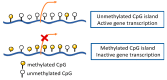
Lung, breast, colorectal, and prostate cancers are the most incident worldwide. Optimal population-based cancer screening methods remain an unmet need, since cancer detection at early stages increases the prospects of successful and curative treatment, leading to a lower incidence of recurrences. Moreover, the current parameters for cancer patients' stratification have been associated with divergent outcomes. Therefore, new biomarkers that could aid in cancer detection and prognosis, preferably detected by minimally invasive methods are of major importance. Aberrant DNA methylation is an early event in cancer development and may be detected in circulating cell-free DNA (ccfDNA), constituting a valuable cancer biomarker. Furthermore, DNA methylation is a stable alteration that can be easily and rapidly quantified by methylation-specific PCR methods. Thus, the main goal of this review is to provide an overview of the most important studies that report methylation biomarkers for the detection and prognosis of the four major cancers after a critical analysis of the available literature. DNA methylation-based biomarkers show promise for cancer detection and management, with some studies describing a "PanCancer" detection approach for the simultaneous detection of several cancer types. Nonetheless, DNA methylation biomarkers still lack large-scale validation, precluding implementation in clinical practice.
Keywords: DNA methylation; biomarker; breast cancer; cell-free DNA; colorectal cancer; detection; liquid biopsy; lung cancer; prognosis; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

