NBCZone: Universal three-dimensional construction of eleven amino acids near the catalytic nucleophile and base in the superfamily of (chymo)trypsin-like serine fold proteases
- PMID: 32151723
- PMCID: PMC7124590
- DOI: 10.1016/j.ijbiomac.2020.03.025
NBCZone: Universal three-dimensional construction of eleven amino acids near the catalytic nucleophile and base in the superfamily of (chymo)trypsin-like serine fold proteases
Abstract
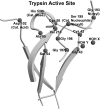
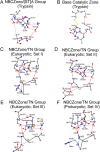
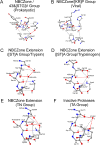
(Chymo)trypsin-like serine fold proteases belong to the serine/cysteine proteases found in eukaryotes, prokaryotes, and viruses. Their catalytic activity is carried out using a triad of amino acids, a nucleophile, a base, and an acid. For this superfamily of proteases, we propose the existence of a universal 3D structure comprising 11 amino acids near the catalytic nucleophile and base - Nucleophile-Base Catalytic Zone (NBCZone). The comparison of NBCZones among 169 eukaryotic, prokaryotic, and viral (chymo)trypsin-like proteases suggested the existence of 15 distinct groups determined by the combination of amino acids located at two "key" structure-functional positions 54T and 55T near the catalytic base His57T. Most eukaryotic and prokaryotic proteases fell into two major groups, [ST]A and TN. Usually, proteases of [ST]A group contain a disulfide bond between cysteines Cys42T and Cys58T of the NBCZone. In contrast, viral proteases were distributed among seven groups, and lack this disulfide bond. Furthermore, only the [ST]A group of eukaryotic proteases contains glycine at position 43T, which is instrumental for activation of these enzymes. In contrast, due to the side chains of residues at position 43T prokaryotic and viral proteases do not have the ability to carry out the structural transition of the eukaryotic zymogen-zyme type.
Keywords: (Chymo)trypsin-like proteases; Catalytic triad; Structural framework; Structural motif.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no conflict of interest.
Figures
Similar articles
-
Structural Catalytic Core in Subtilisin-like Proteins and Its Comparison to Trypsin-like Serine Proteases and Alpha/Beta-Hydrolases.Int J Mol Sci. 2024 Nov 5;25(22):11858. doi: 10.3390/ijms252211858. Int J Mol Sci. 2024. PMID: 39595929 Free PMC article. Review.
-
Structural leitmotif and functional variations of the structural catalytic core in (chymo)trypsin-like serine/cysteine fold proteinases.Int J Biol Macromol. 2021 May 15;179:601-609. doi: 10.1016/j.ijbiomac.2021.03.042. Epub 2021 Mar 10. Int J Biol Macromol. 2021. PMID: 33713772
-
Papain-like cysteine proteinase zone (PCP-zone) and PCP structural catalytic core (PCP-SCC) of enzymes with cysteine proteinase fold.Int J Biol Macromol. 2020 Dec 15;165(Pt A):1438-1446. doi: 10.1016/j.ijbiomac.2020.10.022. Epub 2020 Oct 12. Int J Biol Macromol. 2020. PMID: 33058970 Free PMC article.
-
The catalytic triad of serine peptidases.Cell Mol Life Sci. 2005 Oct;62(19-20):2161-72. doi: 10.1007/s00018-005-5160-x. Cell Mol Life Sci. 2005. PMID: 16003488 Free PMC article. Review.
-
Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications.Proc Natl Acad Sci U S A. 1988 Nov;85(21):7872-6. doi: 10.1073/pnas.85.21.7872. Proc Natl Acad Sci U S A. 1988. PMID: 3186696 Free PMC article.
Cited by
-
Structural Catalytic Core in Subtilisin-like Proteins and Its Comparison to Trypsin-like Serine Proteases and Alpha/Beta-Hydrolases.Int J Mol Sci. 2024 Nov 5;25(22):11858. doi: 10.3390/ijms252211858. Int J Mol Sci. 2024. PMID: 39595929 Free PMC article. Review.
-
The active site of the SGNH hydrolase-like fold proteins: Nucleophile-oxyanion (Nuc-Oxy) and Acid-Base zones.Curr Res Struct Biol. 2023 Dec 29;7:100123. doi: 10.1016/j.crstbi.2023.100123. eCollection 2024. Curr Res Struct Biol. 2023. PMID: 38235349 Free PMC article.
-
Alpha and Omega Classification of β-Lactamase/Transpeptidase-like Superfamily Proteins Based on the Comparison of Their Structural Catalytic Cores.Molecules. 2025 Apr 30;30(9):2019. doi: 10.3390/molecules30092019. Molecules. 2025. PMID: 40363824 Free PMC article.
-
Structural Catalytic Core of the Members of the Superfamily of Acid Proteases.Molecules. 2024 Jul 23;29(15):3451. doi: 10.3390/molecules29153451. Molecules. 2024. PMID: 39124857 Free PMC article. Review.
-
Structural and functional significance of the amino acid differences Val35Thr, Ser46Ala, Asn65Ser, and Ala94Ser in 3C-like proteinases from SARS-CoV-2 and SARS-CoV.Int J Biol Macromol. 2021 Dec 15;193(Pt B):2113-2120. doi: 10.1016/j.ijbiomac.2021.11.043. Epub 2021 Nov 11. Int J Biol Macromol. 2021. PMID: 34774600 Free PMC article.
References
-
- Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247(4):536–540. - PubMed
-
- Dimitriou P.S., Denesyuk A., Takahashi S., Yamashita S., Johnson M.S., Nakayama T., Denessiouk K. Alpha/beta-hydrolases: a unique structural motif coordinates catalytic acid residue in 40 protein fold families. Proteins. 2017;85(10):1845–1855. - PubMed
-
- Nardini M., Dijkstra B.W. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 1999;9(6):732–737. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

