Evaluation of haplotype callers for next-generation sequencing of viruses
- PMID: 32151775
- PMCID: PMC7293574
- DOI: 10.1016/j.meegid.2020.104277
Evaluation of haplotype callers for next-generation sequencing of viruses
Abstract
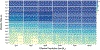
Currently, the standard practice for assembling next-generation sequencing (NGS) reads of viral genomes is to summarize thousands of individual short reads into a single consensus sequence, thus confounding useful intra-host diversity information for molecular phylodynamic inference. It is hypothesized that a few viral strains may dominate the intra-host genetic diversity with a variety of lower frequency strains comprising the rest of the population. Several software tools currently exist to convert NGS sequence variants into haplotypes. Previous benchmarks of viral haplotype reconstruction programs used simulation scenarios that are useful from a mathematical perspective but do not reflect viral evolution and epidemiology. Here, we tested twelve NGS haplotype reconstruction methods using viral populations simulated under realistic evolutionary dynamics. We simulated coalescent-based populations that spanned known levels of viral genetic diversity, including mutation rates, sample size and effective population size, to test the limits of the haplotype reconstruction methods and to ensure coverage of predicted intra-host viral diversity levels (especially HIV-1). All twelve investigated haplotype callers showed variable performance and produced drastically different results that were mainly driven by differences in mutation rate and, to a lesser extent, in effective population size. Most methods were able to accurately reconstruct haplotypes when genetic diversity was low. However, under higher levels of diversity (e.g., those seen intra-host HIV-1 infections), haplotype reconstruction quality was highly variable and, on average, poor. All haplotype reconstruction tools, except QuasiRecomb and ShoRAH, greatly underestimated intra-host diversity and the true number of haplotypes. PredictHaplo outperformed, in regard to highest precision, recall, and lowest UniFrac distance values, the other haplotype reconstruction tools followed by CliqueSNV, which, given more computational time, may have outperformed PredictHaplo. Here, we present an extensive comparison of available viral haplotype reconstruction tools and provide insights for future improvements in haplotype reconstruction tools using both short-read and long-read technologies.
Keywords: Fast-evolving viruses; HIV; Haplotype reconstruction; Intra-host diversity; Next-generation sequencing; Simulations.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no competing interests.
Figures
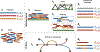

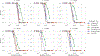
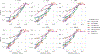


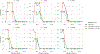
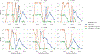
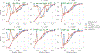

Similar articles
-
Full-length haplotype reconstruction to infer the structure of heterogeneous virus populations.Nucleic Acids Res. 2014 Aug;42(14):e115. doi: 10.1093/nar/gku537. Epub 2014 Jun 27. Nucleic Acids Res. 2014. PMID: 24972832 Free PMC article.
-
Validation of Variant Assembly Using HAPHPIPE with Next-Generation Sequence Data from Viruses.Viruses. 2020 Jul 14;12(7):758. doi: 10.3390/v12070758. Viruses. 2020. PMID: 32674515 Free PMC article.
-
A binning tool to reconstruct viral haplotypes from assembled contigs.BMC Bioinformatics. 2019 Nov 4;20(1):544. doi: 10.1186/s12859-019-3138-1. BMC Bioinformatics. 2019. PMID: 31684876 Free PMC article.
-
Benchmarking of viral haplotype reconstruction programmes: an overview of the capacities and limitations of currently available programmes.Brief Bioinform. 2014 May;15(3):431-42. doi: 10.1093/bib/bbs081. Epub 2012 Dec 19. Brief Bioinform. 2014. PMID: 23257116 Review.
-
Algorithms for Short-Read Viral Haplotype Reconstruction: Challenges, Solutions, and Perspectives.Methods Mol Biol. 2025;2955:89-109. doi: 10.1007/978-1-0716-4702-8_6. Methods Mol Biol. 2025. PMID: 40736895 Review.
Cited by
-
VirStrain: a strain identification tool for RNA viruses.Genome Biol. 2022 Jan 31;23(1):38. doi: 10.1186/s13059-022-02609-x. Genome Biol. 2022. PMID: 35101081 Free PMC article.
-
Novel variants underlying autosomal recessive neurodevelopmental disorders with intellectual disability in Iranian consanguineous families.J Clin Lab Anal. 2022 Feb;36(2):e24241. doi: 10.1002/jcla.24241. Epub 2022 Jan 12. J Clin Lab Anal. 2022. PMID: 35019165 Free PMC article.
-
V-pipe 3.0: a sustainable pipeline for within-sample viral genetic diversity estimation.Gigascience. 2024 Jan 2;13:giae065. doi: 10.1093/gigascience/giae065. Gigascience. 2024. PMID: 39347649 Free PMC article.
-
VILOCA: sequencing quality-aware viral haplotype reconstruction and mutation calling for short-read and long-read data.NAR Genom Bioinform. 2024 Nov 28;6(4):lqae152. doi: 10.1093/nargab/lqae152. eCollection 2024 Dec. NAR Genom Bioinform. 2024. PMID: 39633724 Free PMC article.
-
Quantifying In-Host Quasispecies Evolution.Int J Mol Sci. 2023 Jan 9;24(2):1301. doi: 10.3390/ijms24021301. Int J Mol Sci. 2023. PMID: 36674827 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

