Sirt4 Modulates Oxidative Metabolism and Sensitivity to Rapamycin Through Species-Dependent Phenotypes in Drosophila mtDNA Haplotypes
- PMID: 32152006
- PMCID: PMC7202034
- DOI: 10.1534/g3.120.401174
Sirt4 Modulates Oxidative Metabolism and Sensitivity to Rapamycin Through Species-Dependent Phenotypes in Drosophila mtDNA Haplotypes
Abstract
The endosymbiotic theory proposes that eukaryotes evolved from the symbiotic relationship between anaerobic (host) and aerobic prokaryotes. Through iterative genetic transfers, the mitochondrial and nuclear genomes coevolved, establishing the mitochondria as the hub of oxidative metabolism. To study this coevolution, we disrupt mitochondrial-nuclear epistatic interactions by using strains that have mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) from evolutionarily divergent species. We undertake a multifaceted approach generating introgressed Drosophila strains containing D. simulans mtDNA and D. melanogaster nDNA with Sirtuin 4 (Sirt4)-knockouts. Sirt4 is a nuclear-encoded enzyme that functions, exclusively within the mitochondria, as a master regulator of oxidative metabolism. We exposed flies to the drug rapamycin in order to eliminate TOR signaling, thereby compromising the cytoplasmic crosstalk between the mitochondria and nucleus. Our results indicate that D. simulans and D. melanogaster mtDNA haplotypes display opposite Sirt4-mediated phenotypes in the regulation of whole-fly oxygen consumption. Moreover, our data reflect that the deletion of Sirt4 rescued the metabolic response to rapamycin among the introgressed strains. We propose that Sirt4 is a suitable candidate for studying the properties of mitochondrial-nuclear epistasis in modulating mitochondrial metabolism.
Keywords: Sirt4; TOR pathway; coevolution mtDNA/nDNA.
Copyright © 2020 Sejour et al.
Figures

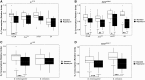


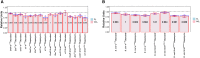

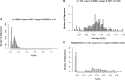
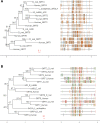
References
-
- Cardamone M. D., Tanasa B., Cederquist C. T., Huang J., Mahdaviani K. et al. , 2018. Mitochondrial Retrograde Signaling in Mammals Is Mediated by the Transcriptional Cofactor GPS2 via Direct Mitochondria-to-Nucleus Translocation. Mol. Cell 69: 757–772.e7. 10.1016/j.molcel.2018.01.037 - DOI - PMC - PubMed
-
- Charif D., and Lobry J. R., 2007. SeqinR 1.0–2: A Contributed Package to the R Project for Statistical Computing Devoted to Biological Sequences Retrieval and Analysis, pp. 207–232 Springer, Berlin, Heidelberg.
-
- Cohen J., 1988. Statistical Power Analysis for the Behavioral Sciences, Ed. 2nd Lawrence Erlbaum Associates, New Jersey.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
