The UCSF Mouse Inventory Database Application, an Open Source Web App for Sharing Mutant Mice Within a Research Community
- PMID: 32152007
- PMCID: PMC7202022
- DOI: 10.1534/g3.120.401086
The UCSF Mouse Inventory Database Application, an Open Source Web App for Sharing Mutant Mice Within a Research Community
Abstract
The UCSF Mouse Inventory Database Application is an open-source Web App that provides information about the mutant alleles, transgenes, and inbred strains maintained by investigators at the university and facilitates sharing of these resources within the university community. The Application is designed to promote collaboration, decrease the costs associated with obtaining genetically-modified mice, and increase access to mouse lines that are difficult to obtain. An inventory of the genetically-modified mice on campus and the investigators who maintain them is compiled from records of purchases from external sources, transfers from researchers within and outside the university, and from data provided by users. These data are verified and augmented with relevant information harvested from public databases, and stored in a succinct, searchable database secured on the university network. Here we describe this resource and provide information about how to implement and maintain such a mouse inventory database application at other institutions.
Keywords: inbred strain; mouse; mutant allele; transgene; web application.
Copyright © 2020 Wall et al.
Figures
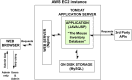
Similar articles
-
Mouse tumor biology database (MTB): enhancements and current status.Nucleic Acids Res. 2000 Jan 1;28(1):112-4. doi: 10.1093/nar/28.1.112. Nucleic Acids Res. 2000. PMID: 10592196 Free PMC article.
-
Using standard nomenclature to adequately name transgenes, knockout gene alleles and any mutation associated to a genetically modified mouse strain.Transgenic Res. 2011 Apr;20(2):435-40. doi: 10.1007/s11248-010-9428-z. Epub 2010 Jul 15. Transgenic Res. 2011. PMID: 20632206
-
Application description and policy model in collaborative environment for sharing of information on epidemiological and clinical research data sets.PLoS One. 2010 Feb 19;5(2):e9314. doi: 10.1371/journal.pone.0009314. PLoS One. 2010. PMID: 20174560 Free PMC article.
-
Annotated expression and activity data for murine recombinase alleles and transgenes: the CrePortal resource.Mamm Genome. 2022 Mar;33(1):55-65. doi: 10.1007/s00335-021-09909-w. Epub 2021 Sep 4. Mamm Genome. 2022. PMID: 34482425 Free PMC article. Review.
-
Murine allele and transgene symbols: ensuring unique, concise, and informative nomenclature.Mamm Genome. 2022 Mar;33(1):108-119. doi: 10.1007/s00335-021-09902-3. Epub 2021 Aug 14. Mamm Genome. 2022. PMID: 34389871 Free PMC article. Review.
