The BIR2/BIR3-Associated Phospholipase Dγ1 Negatively Regulates Plant Immunity
- PMID: 32152212
- PMCID: PMC7210654
- DOI: 10.1104/pp.19.01292
The BIR2/BIR3-Associated Phospholipase Dγ1 Negatively Regulates Plant Immunity
Abstract
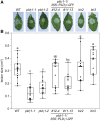
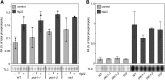
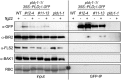
Plants have evolved effective strategies to defend themselves against pathogen invasion. Starting from the plasma membrane with the recognition of microbe-associated molecular patterns (MAMPs) via pattern recognition receptors, internal cellular signaling pathways are induced to ultimately fend off the attack. Phospholipase D (PLD) hydrolyzes membrane phospholipids to produce phosphatidic acid (PA), which has been proposed to play a second messenger role in immunity. The Arabidopsis (Arabidopsis thaliana) PLD family consists of 12 members, and for some of these, a specific function in resistance toward a subset of pathogens has been shown. We demonstrate here that Arabidopsis PLDγ1, but not its close homologs PLDγ2 and PLDγ3, is specifically involved in plant immunity. Genetic inactivation of PLDγ1 resulted in increased resistance toward the virulent bacterium Pseudomonas syringae pv. tomato DC3000 and the necrotrophic fungus Botrytis cinerea As pldγ1 mutant plants responded with elevated levels of reactive oxygen species to MAMP treatment, a negative regulatory function for this PLD isoform is proposed. Importantly, PA levels in pldγ1 mutants were not affected compared to stressed wild-type plants, suggesting that alterations in PA levels are not likely the cause for the enhanced immunity in the pldγ1 line. Instead, the plasma-membrane-attached PLDγ1 protein colocalized and associated with the BAK1-INTERACTING RECEPTOR-LIKE KINASES BIR2 and BIR3, which are known negative regulators of pattern-triggered immunity. Moreover, complex formation of PLDγ1 and BIR2 was further promoted upon MAMP treatment. Hence, we propose that PLDγ1 acts as a negative regulator of plant immune responses in complex with immunity-related proteins BIR2 and BIR3.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures




Comment in
-
A Novel Role for a Phospholipase D in Plant Immunity.Plant Physiol. 2020 May;183(1):33-34. doi: 10.1104/pp.20.00352. Plant Physiol. 2020. PMID: 32385184 Free PMC article. No abstract available.
Similar articles
-
Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens.New Phytol. 2013 Jul;199(1):228-240. doi: 10.1111/nph.12256. Epub 2013 Apr 12. New Phytol. 2013. PMID: 23577648 Free PMC article.
-
BRASSINOSTEROID-SIGNALLING KINASES 7 and 8 associate with the FLS2 immune receptor and are required for flg22-induced PTI responses.Mol Plant Pathol. 2021 Jul;22(7):786-799. doi: 10.1111/mpp.13062. Epub 2021 May 6. Mol Plant Pathol. 2021. PMID: 33955635 Free PMC article.
-
The Arabidopsis Malectin-Like/LRR-RLK IOS1 Is Critical for BAK1-Dependent and BAK1-Independent Pattern-Triggered Immunity.Plant Cell. 2016 Jul;28(7):1701-21. doi: 10.1105/tpc.16.00313. Epub 2016 Jun 17. Plant Cell. 2016. PMID: 27317676 Free PMC article.
-
Responses of Arabidopsis thaliana to challenge by Pseudomonas syringae.Mol Cells. 2008 May 31;25(3):323-31. Epub 2008 May 16. Mol Cells. 2008. PMID: 18483469 Review.
-
The Multifaceted Ubiquitination of BIK1 During Plant Immunity in Arabidopsis thaliana.Int J Mol Sci. 2024 Nov 13;25(22):12187. doi: 10.3390/ijms252212187. Int J Mol Sci. 2024. PMID: 39596247 Free PMC article. Review.
Cited by
-
A Novel Role for a Phospholipase D in Plant Immunity.Plant Physiol. 2020 May;183(1):33-34. doi: 10.1104/pp.20.00352. Plant Physiol. 2020. PMID: 32385184 Free PMC article. No abstract available.
-
Convergent evolution of plant pattern recognition receptors sensing cysteine-rich patterns from three microbial kingdoms.Nat Commun. 2023 Jun 19;14(1):3621. doi: 10.1038/s41467-023-39208-8. Nat Commun. 2023. PMID: 37336953 Free PMC article.
-
Speaking the language of lipids: the cross-talk between plants and pathogens in defence and disease.Cell Mol Life Sci. 2021 May;78(9):4399-4415. doi: 10.1007/s00018-021-03791-0. Epub 2021 Feb 27. Cell Mol Life Sci. 2021. PMID: 33638652 Free PMC article. Review.
-
Dual phosphorylation of DGK5-mediated PA burst regulates ROS in plant immunity.Cell. 2024 Feb 1;187(3):609-623.e21. doi: 10.1016/j.cell.2023.12.030. Epub 2024 Jan 19. Cell. 2024. PMID: 38244548 Free PMC article.
-
The functions of phospholipases and their hydrolysis products in plant growth, development and stress responses.Prog Lipid Res. 2022 Apr;86:101158. doi: 10.1016/j.plipres.2022.101158. Epub 2022 Feb 5. Prog Lipid Res. 2022. PMID: 35134459 Free PMC article. Review.
References
-
- Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerström M(2006) Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J 47: 947–959 - PubMed
-
- Arisz SA, Testerink C, Munnik T(2009) Plant PA signaling via diacylglycerol kinase. Biochim Biophys Acta 1791: 869–875 - PubMed
-
- Bargmann BO, Laxalt AM, Riet BT, Schouten E, van Leeuwen W, Dekker HL, de Koster CG, Haring MA, Munnik T(2006) LePLDβ1 activation and relocalization in suspension-cultured tomato cells treated with xylanase. Plant J 45: 358–368 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

