TEX264 coordinates p97- and SPRTN-mediated resolution of topoisomerase 1-DNA adducts
- PMID: 32152270
- PMCID: PMC7062751
- DOI: 10.1038/s41467-020-15000-w
TEX264 coordinates p97- and SPRTN-mediated resolution of topoisomerase 1-DNA adducts
Abstract
Eukaryotic topoisomerase 1 (TOP1) regulates DNA topology to ensure efficient DNA replication and transcription. TOP1 is also a major driver of endogenous genome instability, particularly when its catalytic intermediate-a covalent TOP1-DNA adduct known as a TOP1 cleavage complex (TOP1cc)-is stabilised. TOP1ccs are highly cytotoxic and a failure to resolve them underlies the pathology of neurological disorders but is also exploited in cancer therapy where TOP1ccs are the target of widely used frontline anti-cancer drugs. A critical enzyme for TOP1cc resolution is the tyrosyl-DNA phosphodiesterase (TDP1), which hydrolyses the bond that links a tyrosine in the active site of TOP1 to a 3' phosphate group on a single-stranded (ss)DNA break. However, TDP1 can only process small peptide fragments from ssDNA ends, raising the question of how the ~90 kDa TOP1 protein is processed upstream of TDP1. Here we find that TEX264 fulfils this role by forming a complex with the p97 ATPase and the SPRTN metalloprotease. We show that TEX264 recognises both unmodified and SUMO1-modifed TOP1 and initiates TOP1cc repair by recruiting p97 and SPRTN. TEX264 localises to the nuclear periphery, associates with DNA replication forks, and counteracts TOP1ccs during DNA replication. Altogether, our study elucidates the existence of a specialised repair complex required for upstream proteolysis of TOP1ccs and their subsequent resolution.
Conflict of interest statement
The authors declare the following competing interests. K.R., J.F., and T.S.M., are inventors of, and Oxford Innovation Ltd. are holders of, a patent application for the use of SPRTN as a cancer biomarker. The remaining authors declare no competiting interests.
Figures
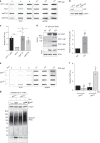

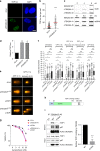
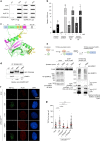
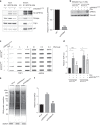
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

