APOE4 is Associated with Differential Regional Vulnerability to Bioenergetic Deficits in Aged APOE Mice
- PMID: 32152337
- PMCID: PMC7062695
- DOI: 10.1038/s41598-020-61142-8
APOE4 is Associated with Differential Regional Vulnerability to Bioenergetic Deficits in Aged APOE Mice
Abstract
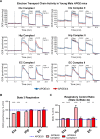
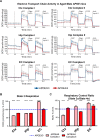
The ε4 allele of apolipoprotein E (APOE) is the dominant genetic risk factor for late-onset Alzheimer's disease (AD). However, the reason for the association between APOE4 and AD remains unclear. While much of the research has focused on the ability of the apoE4 protein to increase the aggregation and decrease the clearance of Aβ, there is also an abundance of data showing that APOE4 negatively impacts many additional processes in the brain, including bioenergetics. In order to gain a more comprehensive understanding of APOE4's role in AD pathogenesis, we performed a transcriptomics analysis of APOE4 vs. APOE3 expression in the entorhinal cortex (EC) and primary visual cortex (PVC) of aged APOE mice. This study revealed EC-specific upregulation of genes related to oxidative phosphorylation (OxPhos). Follow-up analysis utilizing the Seahorse platform showed decreased mitochondrial respiration with age in the hippocampus and cortex of APOE4 vs. APOE3 mice, but not in the EC of these mice. Additional studies, as well as the original transcriptomics data, suggest that multiple bioenergetic pathways are differentially regulated by APOE4 expression in the EC of aged APOE mice in order to increase the mitochondrial coupling efficiency in this region. Given the importance of the EC as one of the first regions to be affected by AD pathology in humans, the observation that the EC is susceptible to differential bioenergetic regulation in response to a metabolic stressor such as APOE4 may point to a causative factor in the pathogenesis of AD.
Conflict of interest statement
K.E.D is on the board of directors of Ceracuity LLC. The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG056387/AG/NIA NIH HHS/United States
- K01 AG061264/AG/NIA NIH HHS/United States
- T32 DK007647/DK/NIDDK NIH HHS/United States
- F32 AG047797/AG/NIA NIH HHS/United States
- K01AG061264/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

