The MHC-II peptidome of pancreatic islets identifies key features of autoimmune peptides
- PMID: 32152506
- PMCID: PMC7117798
- DOI: 10.1038/s41590-020-0623-7
The MHC-II peptidome of pancreatic islets identifies key features of autoimmune peptides
Erratum in
-
Publisher Correction: The MHC-II peptidome of pancreatic islets identifies key features of autoimmune peptides.Nat Immunol. 2020 May;21(5):589. doi: 10.1038/s41590-020-0670-0. Nat Immunol. 2020. PMID: 32238948
Abstract
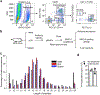
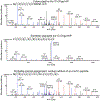
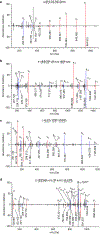
The nature of autoantigens that trigger autoimmune diseases has been much discussed, but direct biochemical identification is lacking for most. Addressing this question demands unbiased examination of the self-peptides displayed by a defined autoimmune major histocompatibility complex class II (MHC-II) molecule. Here, we examined the immunopeptidome of the pancreatic islets in non-obese diabetic mice, which spontaneously develop autoimmune diabetes based on the I-Ag7 variant of MHC-II. The relevant peptides that induced pathogenic CD4+ T cells at the initiation of diabetes derived from proinsulin. These peptides were also found in the MHC-II peptidome of the pancreatic lymph nodes and spleen. The proinsulin-derived peptides followed a trajectory from their generation and exocytosis in β cells to uptake and presentation in islets and peripheral sites. Such a pathway generated conventional epitopes but also resulted in the presentation of post-translationally modified peptides, including deamidated sequences. These analyses reveal the key features of a restricted component in the self-MHC-II peptidome that caused autoreactivity.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




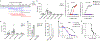

References
-
- Hattori M et al. The NOD mouse: recessive diabetogenic gene in the major histocompatibility complex. Science 231, 733–735 (1986). - PubMed
-
- Todd JA, Bell JI & McDevitt HO HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329, 599–604 (1987). - PubMed
-
- Miyazaki T et al. Direct evidence for the contribution of the unique I-ANOD to the development of insulitis in non-obese diabetic mice. Nature 345, 722–724 (1990). - PubMed
-
- Corper AL et al. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science 288, 505–511 (2000). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

