Omp2b Porin Alteration in the Course of Evolution of Brucella spp
- PMID: 32153552
- PMCID: PMC7050475
- DOI: 10.3389/fmicb.2020.00284
Omp2b Porin Alteration in the Course of Evolution of Brucella spp
Abstract
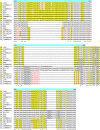
The genus Brucella comprises major pathogenic species causing disease in livestock and humans, e.g. B. melitensis. In the past few years, the genus has been significantly expanded by the discovery of phylogenetically more distant lineages comprising strains from diverse wildlife animal species, including amphibians and fish. The strains represent several potential new species, with B. inopinata as solely named representative. Being genetically more distant between each other, relative to the "classical" Brucella species, they present distinct atypical phenotypes and surface antigens. Among surface protein antigens, the Omp2a and Omp2b porins display the highest diversity in the classical Brucella species. The genes coding for these proteins are closely linked in the Brucella genome and oriented in opposite directions. They share between 85 and 100% sequence identity depending on the Brucella species, biovar, or genotype. Only the omp2b gene copy has been shown to be expressed and genetic variation is extensively generated by gene conversion between the two copies. In this study, we analyzed the omp2 loci of the non-classical Brucella spp. Starting from two distinct ancestral genes, represented by Australian rodent strains and B. inopinata, a stepwise nucleotide reduction was observed in the omp2b gene copy. It consisted of a first reduction affecting the region encoding the surface L5 loop of the porin, previously shown to be critical in sugar permeability, followed by a nucleotide reduction in the surface L8 loop-encoding region. It resulted in a final omp2b gene size shared between two distinct clades of non-classical Brucella spp. (African bullfrog isolates) and the group of classical Brucella species. Further evolution led to complete homogenization of both omp2 gene copies in some Brucella species such as B. vulpis or B. papionis. The stepwise omp2b deletions seemed to be generated through recombination with the respective omp2a gene copy, presenting a conserved size among Brucella spp., and may involve short direct DNA repeats. Successive Omp2b porin alteration correlated with increasing porin permeability in the course of evolution of Brucella spp. They possibly have adapted their porin to survive environmental conditions encountered and to reach their final status as intracellular pathogen.
Keywords: Brucella; Omp2 porin; diversity; evolution; gene conversion; loop.
Copyright © 2020 Cloeckaert, Vergnaud and Zygmunt.
Figures



References
-
- Al Dahouk S., Köhler S., Occhialini A., Jiménez de Bagüés M. P., Hammerl J. A., Eisenberg T., et al. (2017). Brucella spp. of amphibians comprise genomically diverse motile strains competent for replication in macrophages and survival in mammalian hosts. Sci. Rep. 7:44420. 10.1038/srep44420 - DOI - PMC - PubMed
-
- Alton G. G., Jones L. M., Angus R. D., Verger J. M. (1988). Techniques for the Brucellosis Laboratory. Paris: INRA.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

