Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences
- PMID: 32153600
- PMCID: PMC7047749
- DOI: 10.3389/fpls.2020.00022
Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences
Erratum in
-
Corrigendum: Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences.Front Plant Sci. 2020 Mar 20;11:322. doi: 10.3389/fpls.2020.00322. eCollection 2020. Front Plant Sci. 2020. PMID: 32265969 Free PMC article.
Abstract
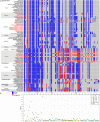
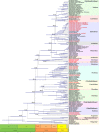
In order to understand the evolution of the orchid plastome, we annotated and compared 124 complete plastomes of Orchidaceae representing all the major lineages in their structures, gene contents, gene rearrangements, and IR contractions/expansions. Forty-two of these plastomes were generated from the corresponding author's laboratory, and 24 plastomes-including nine genera (Amitostigma, Bulbophyllum, Dactylorhiza, Dipodium, Galearis, Gymnadenia, Hetaeria, Oreorchis, and Sedirea)-are new in this study. All orchid plastomes, except Aphyllorchis montana, Epipogium aphyllum, and Gastrodia elata, have a quadripartite structure consisting of a large single copy (LSC), two inverted repeats (IRs), and a small single copy (SSC) region. The IR region was completely lost in the A. montana and G. elata plastomes. The SSC is lost in the E. aphyllum plastome. The smallest plastome size was 19,047 bp, in E. roseum, and the largest plastome size was 178,131 bp, in Cypripedium formosanum. The small plastome sizes are primarily the result of gene losses associated with mycoheterotrophic habitats, while the large plastome sizes are due to the expansion of noncoding regions. The minimal number of common genes among orchid plastomes to maintain minimal plastome activity was 15, including the three subunits of rpl (14, 16, and 36), seven subunits of rps (2, 3, 4, 7, 8, 11, and 14), three subunits of rrn (5, 16, and 23), trnC-GCA, and clpP genes. Three stages of gene loss were observed among the orchid plastomes. The first was ndh gene loss, which is widespread in Apostasioideae, Vanilloideae, Cypripedioideae, and Epidendroideae, but rare in the Orchidoideae. The second stage was the loss of photosynthetic genes (atp, pet, psa, and psb) and rpo gene subunits, which are restricted to Aphyllorchis, Hetaeria, Hexalectris, and some species of Corallorhiza and Neottia. The third stage was gene loss related to prokaryotic gene expression (rpl, rps, trn, and others), which was observed in Epipogium, Gastrodia, Lecanorchis, and Rhizanthella. In addition, an intermediate stage between the second and third stage was observed in Cyrtosia (Vanilloideae). The majority of intron losses are associated with the loss of their corresponding genes. In some orchid taxa, however, introns have been lost in rpl16, rps16, and clpP(2) without their corresponding gene being lost. A total of 104 gene rearrangements were counted when comparing 116 orchid plastomes. Among them, many were concentrated near the IRa/b-SSC junction area. The plastome phylogeny of 124 orchid species confirmed the relationship of {Apostasioideae [Vanilloideae (Cypripedioideae (Orchidoideae, Epidendroideae))]} at the subfamily level and the phylogenetic relationships of 17 tribes were also established. Molecular clock analysis based on the whole plastome sequences suggested that Orchidaceae diverged from its sister family 99.2 mya, and the estimated divergence times of five subfamilies are as follows: Apostasioideae (79.91 mya), Vanilloideae (69.84 mya), Cypripedioideae (64.97 mya), Orchidoideae (59.16 mya), and Epidendroideae (59.16 mya). We also released the first nuclear ribosomal (nr) DNA unit (18S-ITS1-5.8S-ITS2-28S-NTS-ETS) sequences for the 42 species of Orchidaceae. Finally, the phylogenetic tree based on the nrDNA unit sequences is compared to the tree based on the 42 identical plastome sequences, and the differences between the two datasets are discussed in this paper.
Keywords: IR contraction/expansion; Orchidaceae; gene loss; genome rearrangement; plastome evolution.
Copyright © 2020 Kim, Jo, Cheon, Joo, Hong, Kwak and Kim.
Figures








References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

